When you double the number of processors used to solve a given problem, you expect the solution time to be cut in half. However, most programmers know from experience that applications tend to reach a point of diminishing returns when increasing the number of processors being used to solve a fixed-size problem.
How efficiently an application can use more processors is called parallel efficiency, and the most common causes for low parallel efficiency are communication and coordination overheads among processors.
NVSHMEM
NVSHMEM is a communication library based on OpenSHMEM and designed specifically for clusters of NVIDIA GPUs. NVSHMEM can significantly reduce communication and coordination overheads by allowing programmers to perform these operations from within CUDA kernels and on CUDA streams.
Today’s scalable applications typically offload computation phases onto the GPU, but still rely on the CPU for communication, for example using the Message Passing Interface (MPI) or OpenSHMEM. Using the CPU for communication can result in a lot of synchronization overhead between the CPU and GPU, kernel launches, underutilization of the GPU during communication phases, and underutilization of the network during compute phases.
Clever application developers can restructure application code to overlap independent compute and communication phases using CUDA streams. However, this can lead to complex and hard to maintain code. Even with advanced software optimizations, the benefits of this approach may still decrease as strong scaling causes the problem size per GPU to decrease.
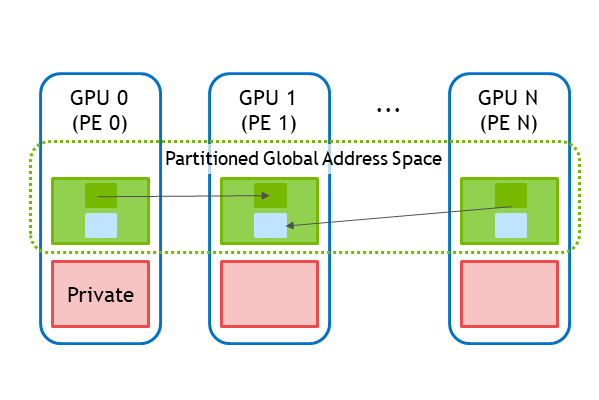
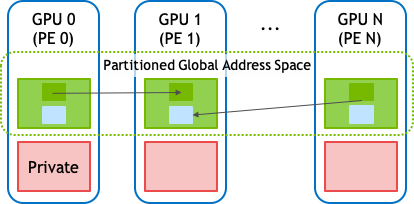
As shown above, NVSHMEM provides a partitioned global address space (PGAS) that combines the memory of multiple GPUs into a shared address space that is accessed through the NVSHMEM API. The NVSHMEM API allows for fine-grained, remote data access that takes advantage of the massive parallelism in the GPU to hide communication overheads.
By performing communication from within CUDA kernels, NVSHMEM allows you to write long running kernels that reduce the amount of overhead generated by synchronization with the CPU. As a result, NVSHMEM can significantly improve parallel efficiency. When needed, NVSHMEM also provides CPU-side calls for inter-GPU communication outside of CUDA kernels and on CUDA streams.
Scalable science with NVSHMEM
To demonstrate the scalability of NVSHMEM, we present three case studies showing how NVSHMEM has been applied to improve the performance of key scientific workloads
- Livermore Big Artificial Neural Network (LBANN)
- Kokkos Conjugate Gradient solver (CGSolve)
- Jacobi method solver: A stencil kernel
Livermore Big Artificial Neural Network
Convolution is a compute-intensive kernel that is used in a wide variety of applications, including image processing, machine learning and scientific computing. The NVIDIA cuDNN library provides a highly optimized single-GPU implementation of convolution for machine learning. Here, we aim to support multi-GPU convolution across a large cluster to accelerate this key operation even further. We use a spatial parallelization approach to scaling convolution, where the spatial domain is broken down into sub-partitions that are distributed to GPUs in the cluster. Halo exchanges that result in nearest-neighbor communications are used to resolve cross-partition data dependencies.
In the Livermore Big Artificial Neural Network (LBANN) deep-learning framework, spatial-parallel convolution is implemented using several different communication methods, including MPI, Aluminum, and NVSHMEM. Both the MPI and Aluminum halo exchanges use CPU-side send and receive operations to exchange data, whereas NVSHMEM uses one-sided put operations.
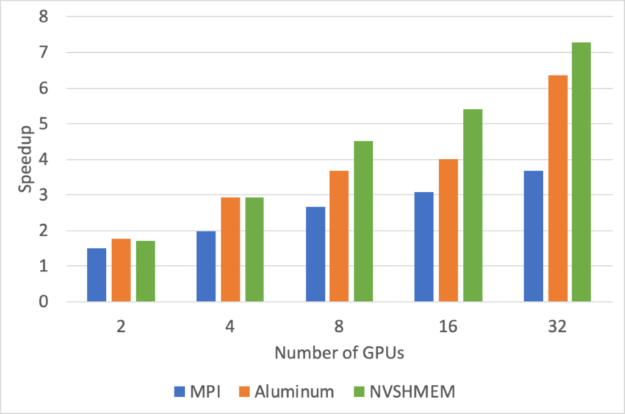
Figure 2 shows the speedups relative to a single GPU for convolution of 1024×1024 images with 16 channels using 16 3×3 filters. All computations are done in single precision.
We used cuDNN version 7.6.5 to perform convolutions and IBM Spectrum MPI 2019.06.24. Performance measurements were done on the Lassen supercomputer at Lawrence Livermore National Laboratory (LLNL), which has two Power 9 CPUs and four Volta GPUs per node. The computation time using just a single GPU is 0.539 ms.
The lower overhead from using NVSHMEM leads to significantly better scaling behavior, versus MPI and Aluminum. At higher GPU counts, these improvements enable nearly twice as much speedup.
The spatial parallel convolution in LBANN is currently available as an experimental feature using the DiHydrogen tensor library. For more information, see LBANN: Livermore Big Artificial Neural Network Toolkit.
Kokkos Conjugate Gradient solver
Kokkos is a parallel programming model that provides performance portability across high performance computing systems. While primarily an on-node programming model, NVSHMEM was recently used to implement an extension of Kokkos to support data structures similar to global arrays.
This removes any need to program communication between GPUs explicitly using MPI and thus significantly reduces code development and tuning effort. You can change the data structure type from local arrays to NVSHMEM backed arrays while otherwise reusing the single GPU code.
The Kokkos Conjugate Gradient solver (CGSolve) implements the conjugate gradient method in Kokkos. CGSolve involves a sequence of kernels that compute the product of two vectors (dot product), perform a sparse matrix-vector multiplication (spmv), and compute a vector sum (axpy). While data could be cleanly partitioned across nodes for the dot product and the vector sum computations, the matrix-vector multiplication accesses vector elements across all partitions. Because of this, it can be challenging to maintain resource utilization when scaling to multiple GPUs. For evaluation, we looked at the following execution scenarios:
- Executions on GPUs within one NVLink complex
- Executions on GPUs within one system node but on different NVLink complexes
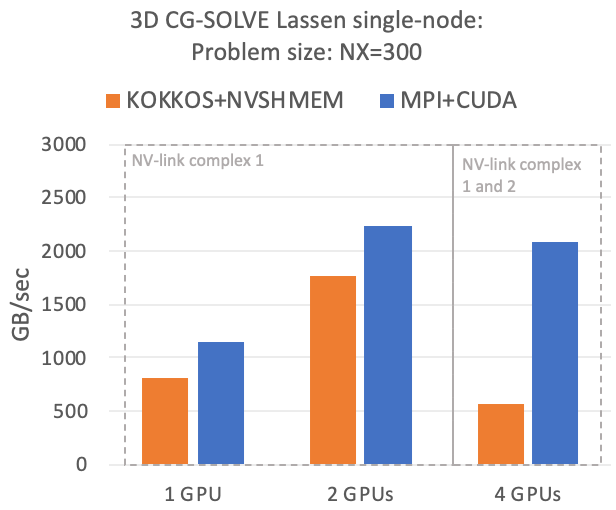
The following figures show a performance baseline for this implementation measured on the Lassen supercomputer at LLNL. Lassen has two IBM Power9 processors per node. Each processor represents a separate NVLink complex that contains two NVIDIA Volta GPUs and connects to one InfiniBand HCA.
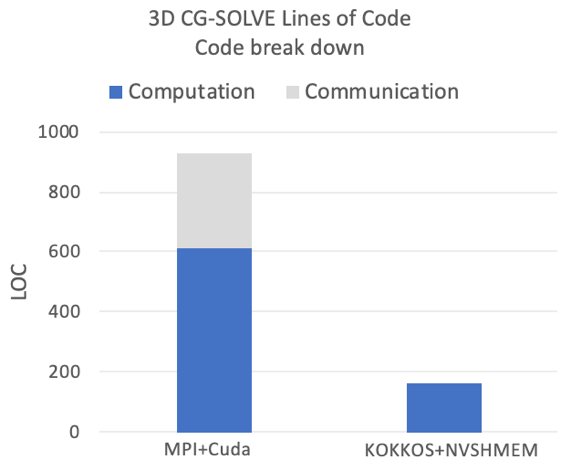
The comparison made in the plots is to a highly tuned MPI+CUDA implementation, which uses complex strategies to overlap communication with calculations. In fact the entire Kokkos+NVSHMEM implementation requires less lines of code than just the pure communication routines of the MPI variant, and the overall code size is almost 6x smaller.
These initial results show that performance with NVSHMEM is competitive within an NVLINK complex; in particular, considering the reduced code complexity, which has significant benefits for maintainability and development cost. When crossing the NVLINK complex, the performance drops compared to the MPI variant. This is caused primarily by high remote access latencies when leaving an NVLINK complex. We are now investigating communication-coarsening optimizations as well as aggregation and caching approaches in the low-level communication layer to close this gap. The code is also functional in an inter-node setting, but more optimization work has to be done to improve the performance.
Resources:
Jacobi method solver: A stencil kernel
The Jacobi method is an iterative approach to solving systems of linear equations. Jacobi is referred to as a stencil computation because the values surrounding a given element in the matrix are needed when computing each element.
We created a Jacobi kernel that uses NVSHMEM to exchange data between iterations and explore its scalability on the Summit supercomputer located at the Oak Ridge Leadership Computing Facility (OLCF). Summit is an IBM AC922 system that links more than 27,000 NVIDIA Volta GPUs with more than 9,000 IBM Power9 CPUs, interconnected with dual-rail Mellanox EDR 100Gb/s InfiniBand. The Summit supercomputer has more than 10 petabytes of memory paired with fast, high-bandwidth pathways for efficient data movement.
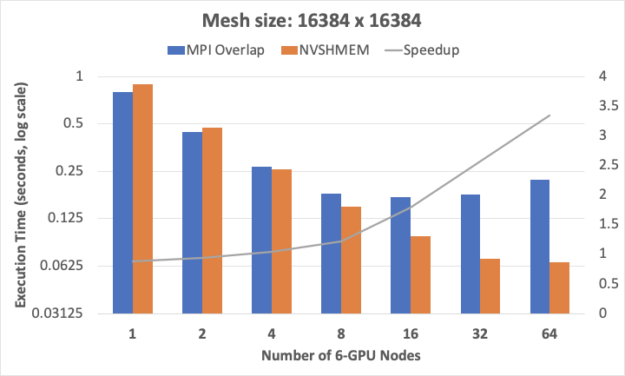
We used a fixed problem size of 16Kx16K, measured the execution time for solutions based on each of MPI and NVSHMEM, and calculated the relative performance advantage of NVSHMEM over MPI as the number of nodes was increased.
The MPI baseline implementation is aggressively optimized to overlap communications. However, benefits from overlap diminish as compute and communication times reduce with less data per node, as would happen in strong scaling. Further, latencies from CPU-GPU synchronization to achieve inter-GPU communication tend to be on the critical path. These latencies form a considerable and increasing fraction of the execution time as we scale to a larger number of GPUs. These synchronization latencies are avoided in the NVSHMEM version by using GPU-initiated communication.
In Figure 4, NVSHMEM’s efficiency advantages over MPI are graphically illustrated with MPI’s 16-node scaling limit while NVSHMEM execution times continue to drop, and with a rising relative speedup advantage for NVSHMEM that is as high as 3.3x on 64 6-GPU nodes.
For more information, see An Initial Assessment of NVSHMEM for High Performance Computing (2020 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW)).
For more information about the code, see the following resources:
Conclusion
NVSHMEM is a new communication API for clusters of NVIDIA GPUs. NVSHMEM creates a global address space that includes the memory of all GPUs in the cluster and provides APIs for GPU-initiated, in-kernel communication. NVSHMEM can provide low communication overheads that improve an application’s strong scaling efficiency.
For more information, see GTC 2020: A Partitioned Global Address Space Library for Large GPU Clusters.
Acknowledgements
This research was funded by the United States Department of Defense and the United States Department of Energy. This work used computing resources of the Oak Ridge Leadership Computing Facility at Oak Ridge National Laboratory, Oak Ridge, Tennessee and the Livermore Computing Center at Lawrence Livermore National Laboratory, Livermore, California.
This work used resources available to Sandia National Laboratories. Sandia National Laboratories is a multimission laboratory managed and operated by National Technology and Engineering Solutions of Sandia, LLC., a wholly owned subsidiary of Honeywell International, Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA-0003525.
Work on the Livermore Big Artificial Neural Network toolkit was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344 (LLNL-WEB-813476).