This post delves into the capabilities of decoding DICOM medical images within AWS HealthImaging using the nvJPEG2000 library. We’ll guide you through the intricacies of image decoding, introduce you to AWS HealthImaging, and explore the advancements enabled by GPU-accelerated decoding solutions.
Embarking on a journey to enhance throughput and reduce costs in deciphering medical images within AWS HealthImaging through the GPU-accelerated nvJPEG2000 library represents a significant stride towards operational efficiency in cloud environments. These innovations promise substantial savings, with projections suggesting potential cost reductions totaling in the hundreds of millions of USD for such workloads.
JPEG 2000
Implementing JPEG 2000 correctly poses significant complexity, with early encounters of interoperability issues hindering seamless integration across different systems. This complexity acts as a barrier to widespread adoption. However, the introduction of the High-Throughput JPEG 2000 (HTJ2K) encoding system marks a notable advancement in image compression technology. HTJ2K, outlined in part 15 of the JPEG 2000 standard, enhances throughput by replacing the original block coding algorithm, EBCOT (Embedded Block Coding with Optimized Truncation), with the more efficient FBCOT (Fast Block Coding with Optimized Truncation).
This new standard addresses the decoding speed limitations and opens the door to a more widespread adoption of JPEG 2000 in the medical imaging domain. HTJ2K accommodates both lossless and lossy compression, offering a balance between preserving critical medical details and achieving efficient storage. Grayscale and color images with arbitrary width and height, along with support for up to 16 bits per channel, showcase the adaptability of HTJ2K. The new standard imposes no restrictions on decomposition levels, enabling a wide range of options.
nvJPEG2000 library
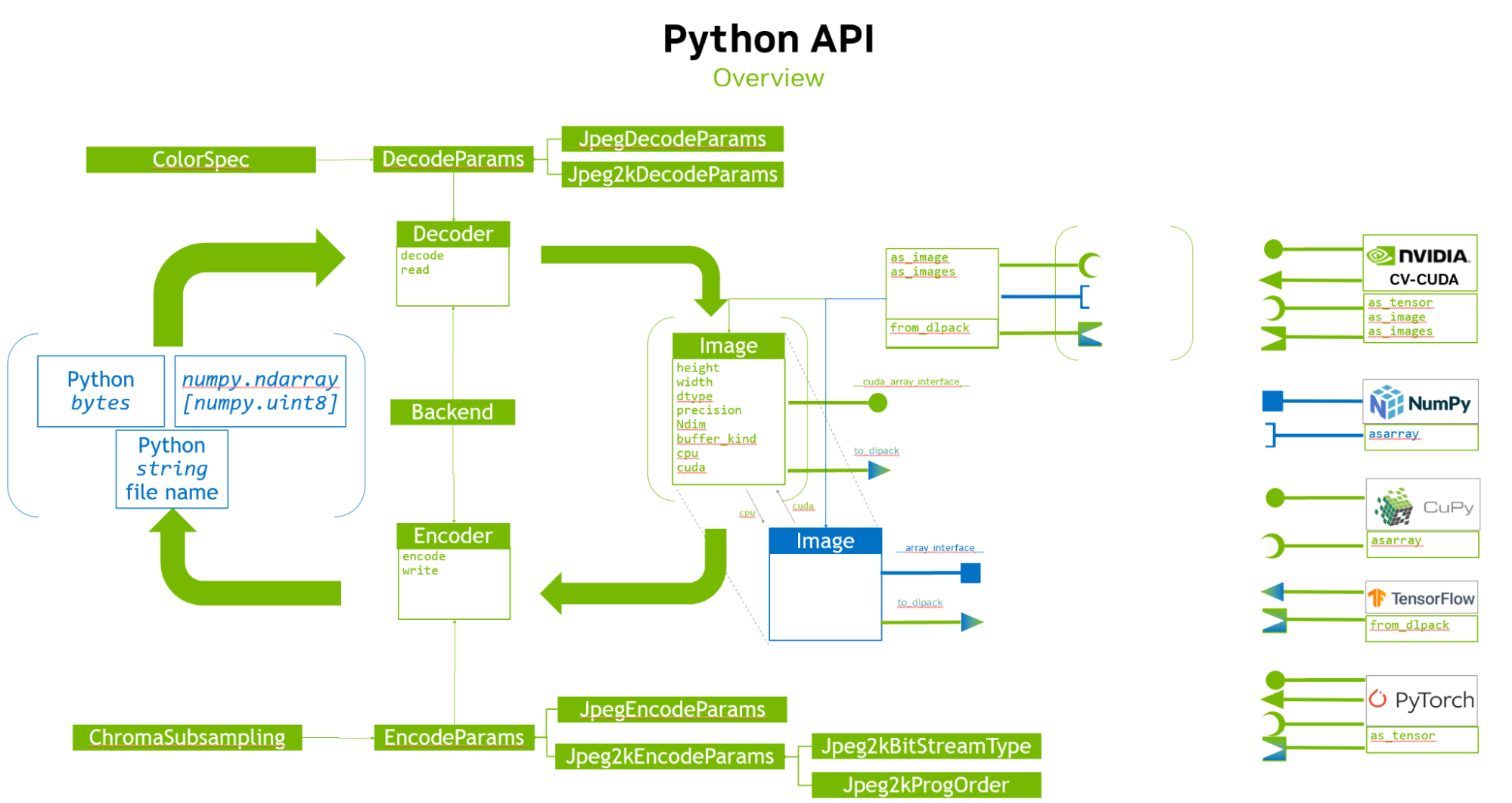
With the advent of GPU acceleration through libraries like nvJPEG2000, the decoding performance of HTJ2K has reached new heights. This advancement unlocks the true potential of JPEG 2000 in medical image processing, making it a viable and efficient solution for healthcare providers, researchers, and developers. nvJPEG2000 provides a C API, including functions like nvjpeg2kDecode for decoding a single image and nvjpeg2kDecodeTile for decoding specific tiles within an image. The library offers:
- Unified API Interface nvImageCodec: The open source library seamlessly integrates with Python, offering a convenient interface for developers.
- Decoding performance analysis: Comparative decoding performance analysis between HTJ2K and traditional JPEG 2000, including insights into GPU acceleration.
To ensure usability, high performance, and readiness for production, this post explores the integration of HTJ2K decoding with MONAI, a framework designed for medical image analysis. MONAI Deploy App SDK provides high-performance capabilities, and facilitation of debugging in medical imaging AI applications. It also provides an in-depth examination of the cost benefits associated with using AWS HealthImaging, MONAI, and nvJPEG2000 for medical image processing.
Enterprise-scale medical image storage with AWS HealthImaging
Thanks to lossless HTJ2K encoding and the AWS high-performance network backbone, AWS HealthImaging offers subsecond image retrieval from anywhere and fast access to images stored in the cloud. It is designed to be workflow-agnostic, seamlessly integrating into existing medical imaging workflows. It is also a DICOM-compliant solution, ensuring interoperability and adherence to industry standards in medical image communication. The service provides native API for scalable and fast image ingestion, accommodating the ever-growing volume of medical imaging data.
GPU-accelerated image decoding
To further enhance the image decoding performance, AWS HealthImaging supports GPU acceleration, specifically leveraging the NVIDIA nvJPEG2000 library. This GPU acceleration ensures rapid and efficient decoding of medical images, enabling healthcare providers to access critical information with unprecedented speed. The supported features of HTJ2K decoding encompass a wide range of options to accommodate diverse image types, sizes, compression needs, and decoding scenarios, making it a versatile and adaptable choice for various image processing applications. These include the following:
- Image types: HTJ2K is compatible with grayscale and color images of arbitrary width and height. It accommodates diverse image formats and dimensions.
- Bit depth: HTJ2K handles images with a bit depth of up to 16 bits per channel. This high bit depth support ensures accurate representation of color and detail.
- Lossless compression: The HTJ2K standard includes support for lossless compression, enabling the preservation of image quality without any data loss.
- Uniform code block configuration: All code blocks within HTJ2K conform to the HT (high throughput) standard. There is no use of refinement code blocks, simplifying the decoding process.
- Code block sizes: HTJ2K employs different code block sizes, including 64×64, 32×32, and 16×16. This adaptability enables efficient representation of images with varying levels of detail and complexity.
- Progression orders: HTJ2K supports multiple progression orders, including those listed below. These progression orders offer flexibility in how the image data is organized and transmitted.
- LRCP (Layer-Resolution-Component-Position)
- RLCP (Resolution-Layer-Component-Position)
- RPCL (Resolution-Position-Component-Layer)
- PCRL (Position-Component-Resolution-Layer)
- CPRL (Component-Position-Resolution-Layer)
- Variable decomposition levels: The standard allows for various numbers of decomposition levels, ranging from 1 to 5. This flexibility in decomposition provides options for optimizing image compression based on specific requirements.
- Multi-tile decoding with different tile sizes: HTJ2K is capable of decoding images divided into multiple tiles, and these tiles can have different sizes. This feature enhances the ability to efficiently decode large images by processing them in smaller, manageable sections.
AWS HealthImaging walkthrough
In this walkthrough, we showcase the use of AWS HealthImaging. We demonstrate the process of employing a SageMaker multimodel endpoint for image decoding, utilizing a GPU-accelerated interface.
Step 1: Stage your DICOM images
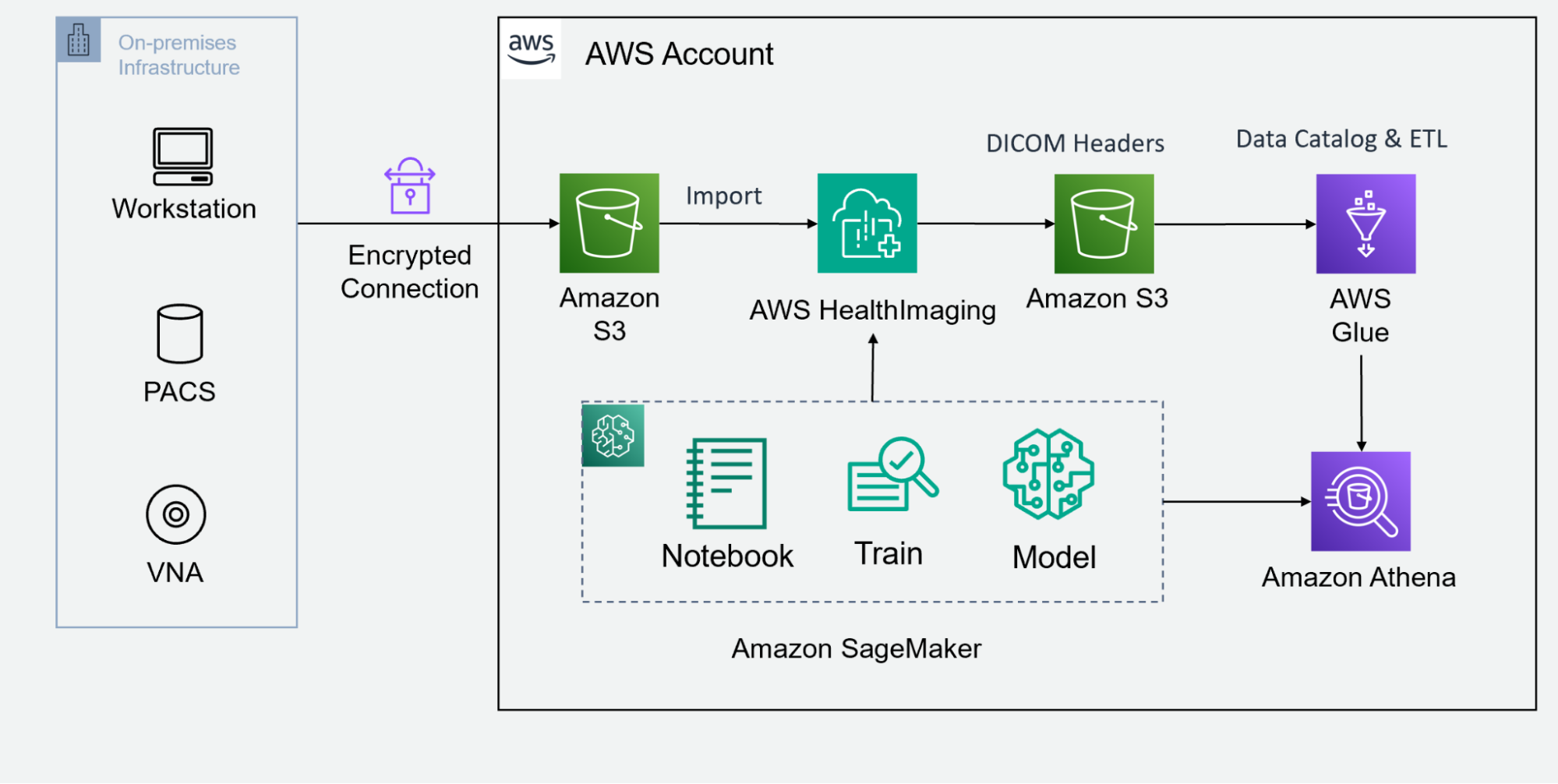
Begin by staging your DICOM images in an Amazon S3 bucket. AWS HealthImaging integrates with launching partner products, offering a variety of tools that best suit your workflow and upload and organize your DICOM image data in the designated S3 bucket. You can explore the AWS Open Data Program as well. There are open datasets with synthetic medical imaging data in public S3 buckets, such as Synthea Coherent.
Step 2: Invoke the API for DICOM data import
Once your DICOM images are staged in the S3 bucket, the next step involves invoking a native API to import the DICOM data into AWS HealthImaging. This managed API facilitates a smooth and automated process, ensuring that your medical image data is efficiently transferred and ready for further optimization.
Step 3: Index DICOM headers in a data lake
After successful import, retrieve DICOM headers from AWS HealthImaging, decompress the data blobs, and write these JSON objects to a data lake S3 bucket. From there, you can leverage the AWS data lake analytics tools, like Amazon Glue to generate a data catalog, Amazon Athena to perform ad hoc SQL query, and Amazon QuickSight to build data visualization dashboard. You can also combine image metadata with other health data modalities to perform multimodal data analytics.
Step 4: Access your medical image data
With managed APIs, access becomes a seamless experience. AWS HealthImaging provides high performant and granular access of your imaging data with subsecond speeds.
AWS partners who build their PACS viewers and VNA solutions on the cloud can integrate their image viewing applications with AWS HealthImaging. These applications are optimized to deliver a user-friendly and efficient experience for viewing and analyzing medical images at scale. Examples of AWS partners PACS include Allina Health case study, Visage Imaging, and Visage AWS.
Scientists and researchers can leverage Amazon SageMaker to perform AI and ML modeling, to unlock advanced insights and automate reviewing and annotation tasks. Amazon Sagemaker can be used with MONAI for developing robust AI models. Using Amazon SageMaker notebook, users can retrieve the pixel frames from AWS HealthImaing, visualize medical images using open source tool like itkwidget, and create SageMaker managed training jobs or model hosting endpoints.
As a HIPAA-eligible service, AWS HealthImaging offers flexibility to grant and audit secure access to the medical image data to remote users. The access control is managed by Amazon Identity and Access Management, which ensures that authorized users can have granular access the ImageSet data. The access activities can also be monitored by Amazon CloudTrail, to track who has accessed which data by what time.
Step 5: GPU-enabled HTJ2K decoding
In a typical AI or ML workflow (CPU decoding pathway), the HTJ2K-encoded pixel frames will be loaded into CPU memory, then decoded and converted to the tensors in the CPU. These can be copied and processed by GPUs. nvJPEG2000 can take the encoded pixels from AWS HealthImaging and decode them into GPU memory directly (GPU decoding pathway), and MONAI has built-in functions to transform image data into tensors that are ready to feed in deep learning models. It is a shorter path compared to the CPU decoding approach, as illustrated in Figure 2.
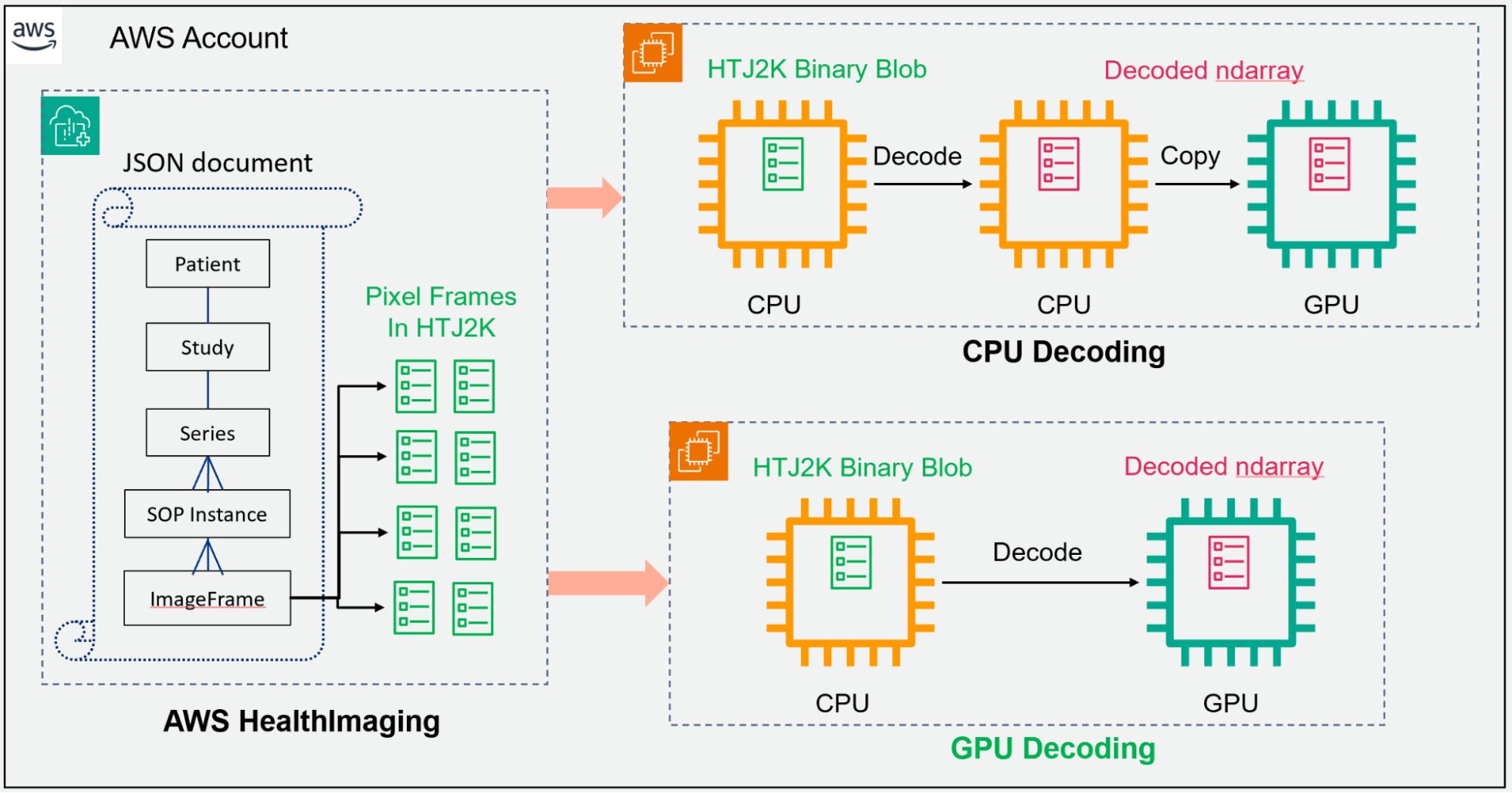
Additionally, GPU acceleration with nvJPEG2000 significantly improves decoding performance, reducing latency and enhancing overall responsiveness. The library seamlessly integrates with Python, providing developers with a familiar and powerful environment for image decoding tasks.
A demo notebook running on Amazon SageMaker showcases how to integrate and leverage the power of GPU-accelerated image decoding in a scalable and efficient manner. In our experiment, the GPU decoding on SageMaker g4dn.2xlarge instance is ~7 times faster than CPU decoding on SageMaker m5.2xlarge instance (Figure 3).
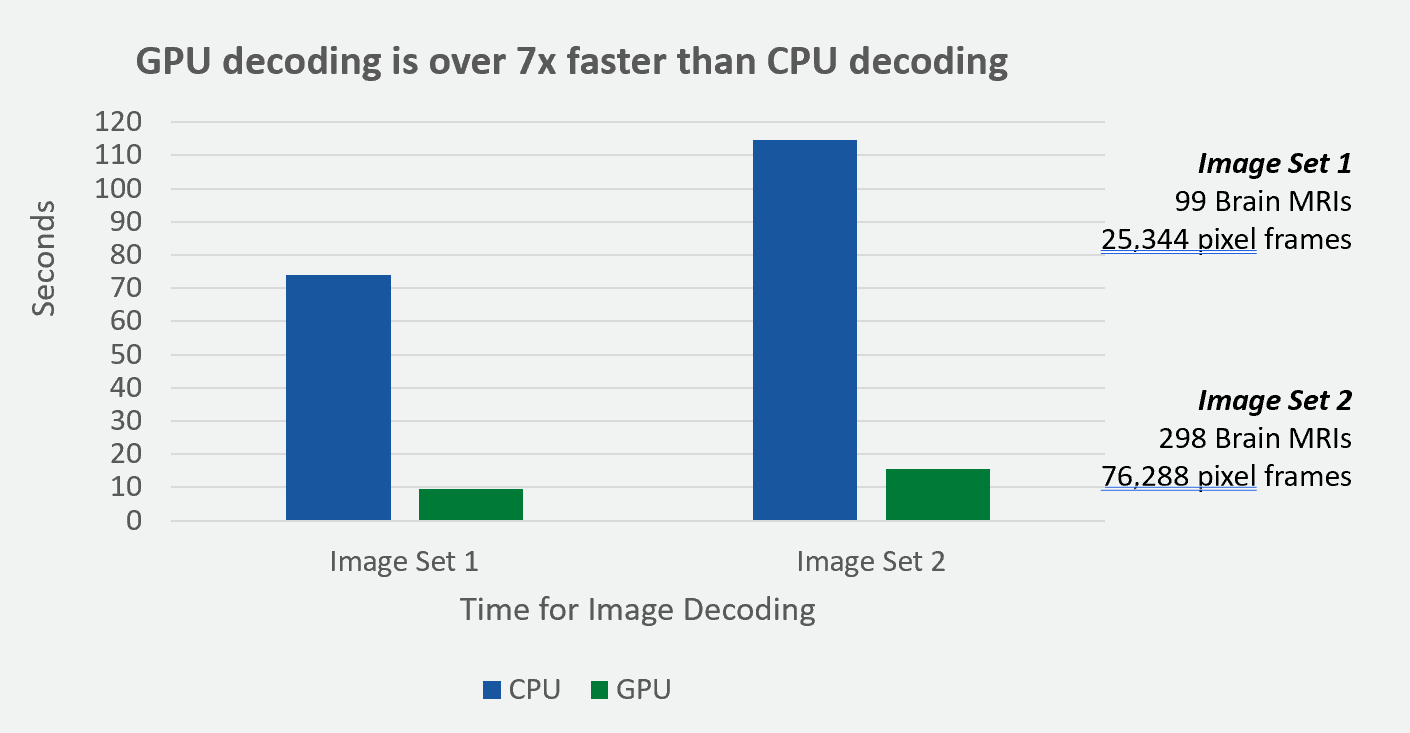
In this experiment, we used the synthetic brain MRI images from the Synthea Coherent dataset. The GPU acceleration demonstrated a similar speed-up factor for different sizes of datasets. The image sets labeled above contain Brain MRIs and pixel frames. These pixel frames represent DICOM MRI images, and they are encoded in the compressed HTJ2K data format.
Cost-benefit analysis
Combined with advanced image decoding technologies, AWS HealthImaging not only improves efficiency but also provides a cost-effective solution for healthcare organizations. The end-to-end cost benefits of the proposed solution are substantial, particularly when considering the impressive throughput speedup achieved with GPU acceleration.
The speedup on a single NVIDIA T4 GPU on EC2 G4 instance surpasses the CPU baseline by approximately 5x, and this improvement is further enhanced on the new L4 GPU on EC2 G6 instance to an impressive 12x. Scaling with multiple GPU instances, the performance exhibits nearly linear scalability, reaching around 19x and 48x on four NVIDIA T4 GPUs and four NVIDIA L4 GPUs, respectively.
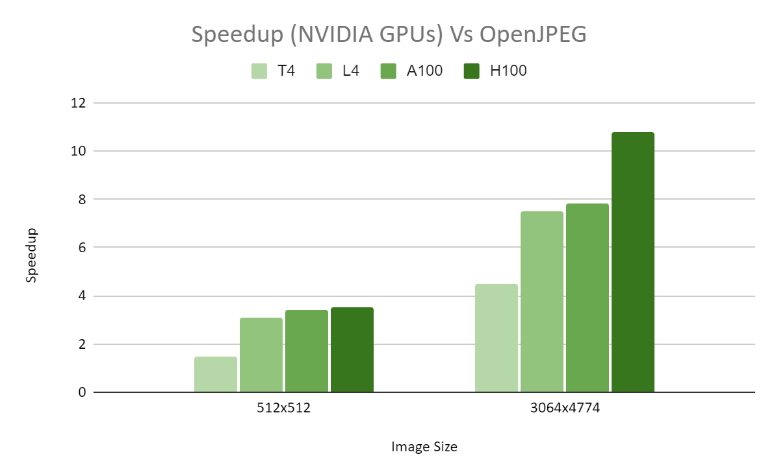
In terms of decoding performance, we carried out a comparative analysis with OpenJPEG. For a CT1 16-bit 512×512 grayscale image, we noticed a significant 2.5x speed enhancement across diverse GPU configurations. Moreover, for the larger MG1 16-bit grayscale image measuring 3064×4774, we accomplished an impressive 8x speed increase across various GPU setups.
For a comprehensive assessment of annual cloud costs and energy usage, our calculations are based on a standard segmentation workload. This workload involves uploading 500 DICOM files per minute to a MONAI server platform. Our cost estimation focuses solely on T4 GPUs at present, with L4 GPUs expected in the future. We assume one-year reserved pricing for Amazon EC2 G4 instances.
Under these conditions, the annual cost of processing the DICOM workload on a single T4 GPU is estimated to be approximately $74 million, compared to the $345.4 million associated with the CPU pipeline. This represents a significant reduction in cloud expenditure, with forecasts indicating potential savings in the hundreds of millions of dollars for such hospital-based workloads.
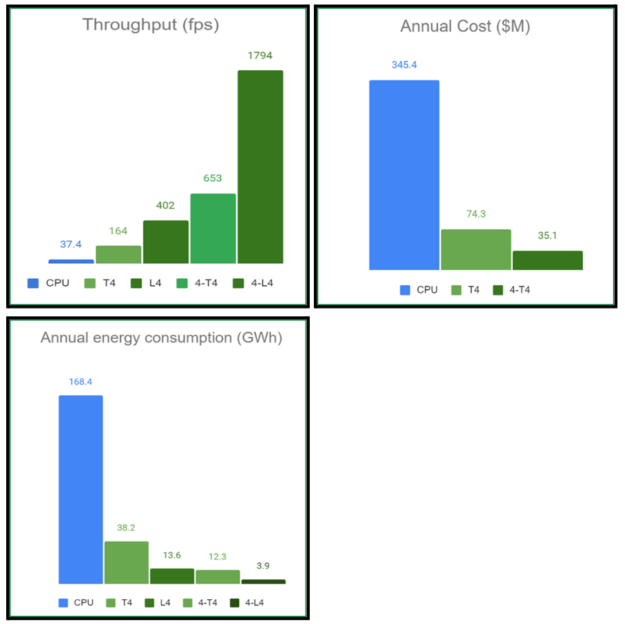
On a single T4 GPU, the end-to-end throughput speedup is approximately 5x faster compared to the CPU baseline. This speedup is further enhanced on the new L4 GPU to around 12x faster. When utilizing multiple GPU instances, the performance scales almost linearly. For example, with four T4 GPUs, the speedup reaches approximately 19x, while with four L4 GPUs, it escalates to approximately 48x faster.
Considering the environmental impact, energy efficiency is a critical factor for data centers handling large workloads. Our calculations reveal that the annual energy consumption, measured in GWh, is considerably reduced when utilizing the respective GPU-based hardware. Specifically, the energy consumption on a single L4 system is approximately one-twelfth that of a CPU server.
For a workload similar to the example DICOM video scenario (500 hours of video per minute), the annual energy savings estimates are on the order of hundreds of GWh. These energy savings are not only economically beneficial, but also environmentally significant. The equivalent reduction in greenhouse gas emissions is substantial, similar to avoiding emissions from tens of thousands of passenger vehicles driven in a year, each covering approximately 11,000 miles annually.
Why nvImageCodec?
The NVIDIA/nvImageCodec library provides developers with a robust and efficient solution for image decoding tasks. Leveraging the power of NVIDIA GPUs, nvImageCodec delivers accelerated decoding performance, making it ideal for applications requiring high throughput and low latency.
Key features
- GPU acceleration: One of the standout features of nvImageCodec is its GPU acceleration capabilities. By harnessing the computational power of NVIDIA GPUs, nvImageCodec significantly speeds up the image decoding process, allowing for faster processing of large datasets.
- Seamless integration: nvImageCodec seamlessly integrates with Python, providing developers with a familiar environment for their image processing workflows. With a user-friendly API, integrating nvImageCodec into existing Python projects is straightforward.
- High performance: With optimized algorithms and parallel processing, nvImageCodec delivers exceptional performance, even when dealing with complex image decoding tasks. Whether you’re decoding JPEG, JPEG 2000, TIFF, or other image formats, nvImageCodec ensures speedy and efficient processing.
- Versatility: From medical imaging to computer vision applications, nvImageCodec supports a wide range of use cases. Whether you’re working with grayscale or color images, nvImageCodec offers versatility and flexibility to meet your image decoding needs.
Use cases
- Medical imaging: In the field of medical imaging, fast and accurate image decoding is crucial for timely diagnosis and treatment. With nvImageCodec, healthcare professionals can decode medical images with speed and precision, enabling faster decision-making and improved patient outcomes.
- Computer vision: In computer vision applications, image decoding speed plays a vital role in real-time processing tasks such as object detection and image classification. By leveraging nvImageCodec’s GPU acceleration, developers can achieve high-performance image decoding, enhancing the responsiveness of their applications.
- Remote sensing: In remote sensing applications, decoding large satellite images quickly and efficiently is essential for various tasks such as environmental monitoring and disaster management. With nvImageCodec, researchers and analysts can decode satellite images with ease, enabling timely analysis and decision-making.
How to get nvImageCodec
Obtaining nvImageCodec is straightforward. You can download it from multiple sources such as PyPI, the NVIDIA Developer Zone, or directly from the GitHub repository. Once downloaded, you can begin experimenting with encoding and decoding examples to enhance the efficiency of your image codec pipeline.
How to batch decode high-throughput JPEG 2000 medical images
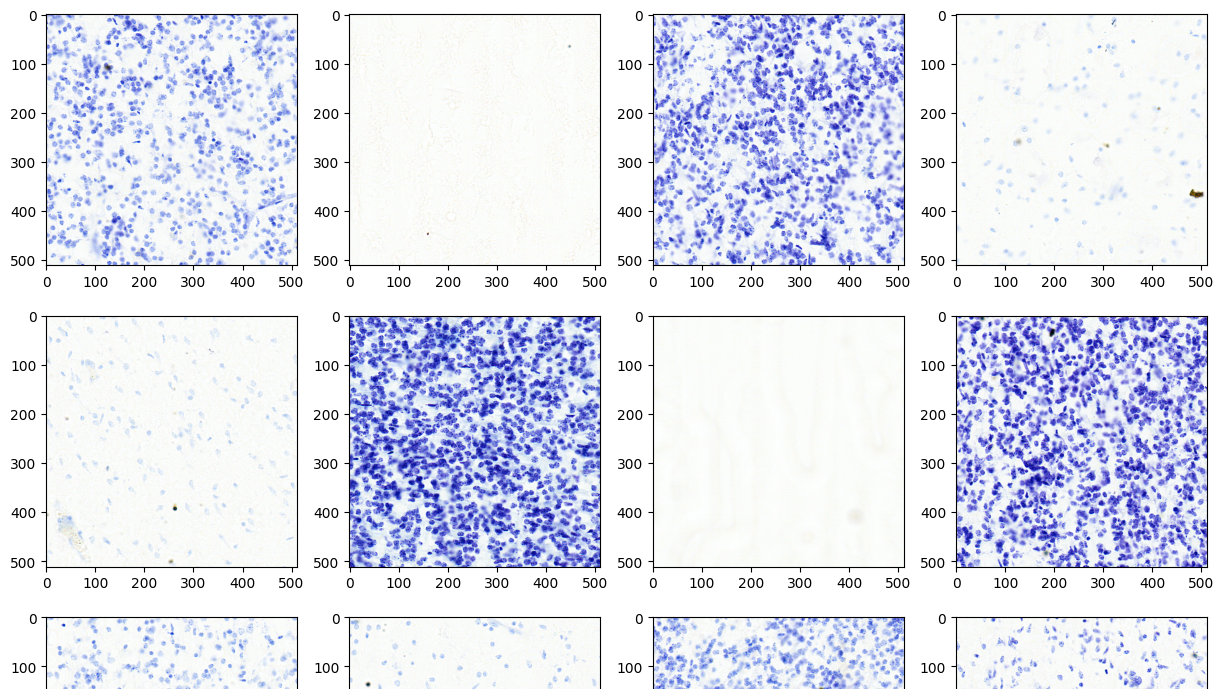
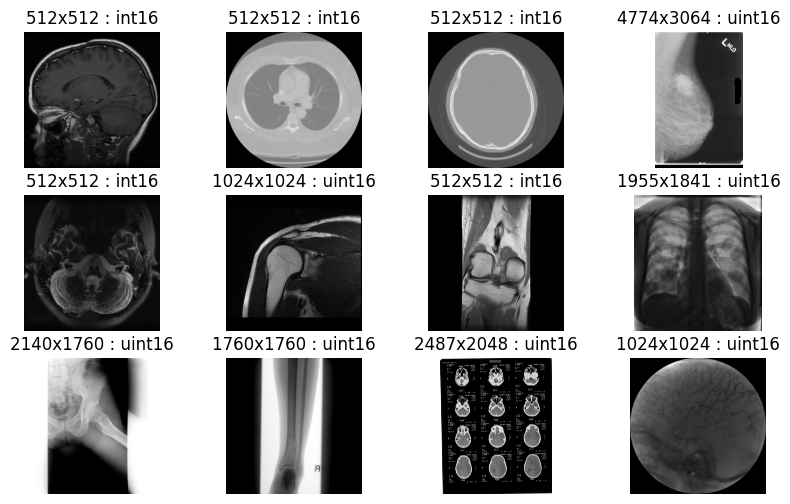
Below is a Python example demonstrating batch image decoding using the nvImageCodec library. This example illustrates how to use the nvImageCodec to decode HTJ2K images in bulk. All images within the specified folder are compressed in lossless HTJ2K format with a precision of uint16 bits. The output confirms successful decoding of all medical images without any loss in quality (Figure 8).
import os; import os.path
from matplotlib import pyplot as plt
from nvidia import nvimgcodec
dir = "htj2k_lossless"
image_paths = [os.path.join(dir, filename) for filename in os.listdir(dir)]
decode_params = nvimgcodec.DecodeParams(allow_any_depth = True, color_spec=nvimgcodec.ColorSpec.UNCHANGED)
nv_imgs = nvimgcodec.Decoder().read(image_paths, decode_params)
cols= 4
rows = (len(nv_imgs)+cols-1)//cols
fig, axes = plt.subplots(rows, cols); fig.set_figheight(2*rows); fig.set_figwidth(10)
for i in range(len(nv_imgs)):
axes[i//cols][i%cols].set_title("%ix%i : %s"%(nv_imgs[i].height, nv_imgs[i].width, nv_imgs[i].dtype));
axes[i//cols][i%cols].set_axis_off()
axes[i//cols][i%cols].imshow(nv_imgs[i].cpu(), cmap='gray')
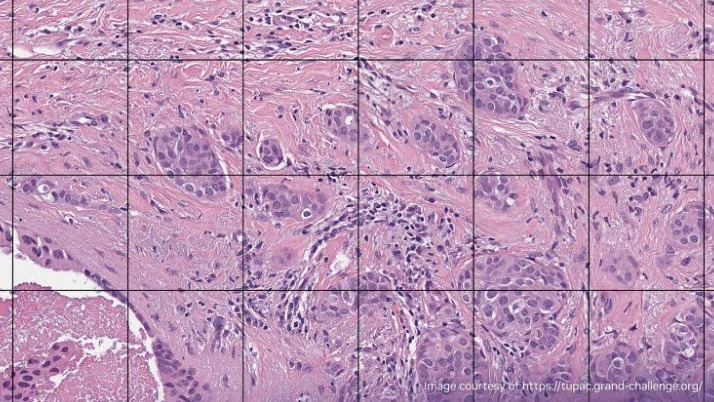
How to batch decode multiple JPEG 2000 tiles
Below is a Python example showcasing tile-based image decoding for large images, employing the nvImageCodec library. This demonstrates the process of using the nvImageCodec to decode JPEG 2000 compressed images of substantial size. Each tile represents a region of interest (ROI), measuring 512 x 512 pixels.
The decoding process involves segmenting the image into tiles, determining the total number of regions, and then using the nvImageCodec to decode individual tiles based on their indices, providing specific tile decoding information. The resulting output displays information pertaining to the different tiles.
from matplotlib import pyplot as plt
import numpy as np
import random; random.seed(654321)
from nvidia import nvimgcodec
jp2_stream = nvimgcodec.CodeStream('./B_37_FB3-SL_570-ST_NISL-SE_1708_lossless.jp2')
def get_region_grid(stream, roi_height, roi_width):
regions = []
num_regions_y = int(np.ceil(stream.height / roi_height))
num_regions_x = int(np.ceil(stream.width / roi_width))
for tile_y in range(num_regions_y):
for tile_x in range(num_regions_x):
tile_start = (tile_y * roi_height, tile_x * roi_width)
tile_end = (np.clip((tile_y + 1) * roi_height, 0, stream.height), np.clip((tile_x + 1) * roi_width, 0, stream.width))
regions.append(nvimgcodec.Region(start=tile_start, end=tile_end))
print(f"{len(regions)} {roi_height}x{roi_width} regions in total")
return regions
regions_native_tiles = get_region_grid(jp2_stream, jp2_stream.tile_height, jp2_stream.tile_width) # 512x512 tiles
dec_srcs = [nvimgcodec.DecodeSource(jp2_stream, region=regions_native_tiles[random.randint(0, len(regions_native_tiles)-1)]) for k in range(16)]
imgs = nvimgcodec.Decoder().decode(dec_srcs)
fig, axes = plt.subplots(4, 4)
fig.set_figheight(15)
fig.set_figwidth(15)
i = 0
for ax0 in axes:
for ax1 in ax0:
ax1.imshow(np.array(imgs[i].cpu()))
i = i + 1
Conclusion
Whether you’re a healthcare provider, researcher, or developer, the resurgence of JPEG 2000 alongside cutting-edge technologies unveils fresh avenues for innovation in the critical domain of medical imaging. AWS HealthImaging, coupled with advanced compression standards and GPU acceleration, emerges as a crucial tool for healthcare professionals striving to enhance diagnostic capabilities and elevate patient outcomes.
This innovation also ushers in novel avenues for high-performance multimodal data analysis, seamlessly integrating genomic, clinical, and medical imaging data to extract meaningful insights. The managed data science platform on the cloud streamlines model training and deployment processes, reducing friction. Embrace these advancements and drive the future of healthcare forward through accelerated and robust image decoding.
To get started, explore nvImageCodec, the MONAI on AWS workshop, and Multimodal Data Analysis with AWS Health and Machine Learning Services. For more information, see:
- Accelerating JPEG 2000 Decoding for Digital Pathology and Satellite Images Using the nvJPEG2000 Library
- Accelerate Your Medical Imaging Research with MONAI on AWS
- Decoding Medical Images in AWS HealthImaging Using NVIDIA nvJPEG2000
- Whole Human Brain Neuro-Mapping at Cellular Resolution on NVIDIA DGX