Whole human brain imaging of 100 brains at a cellular level within a 2-year timespan, and subsequent analysis and mapping, requires accelerated supercomputing and computational tools. This need is well matched by NVIDIA technologies, which range across hardware, computational systems, high-bandwidth interconnects, domain-specific libraries, accelerated toolboxes, curated deep-learning models, and container runtimes. NVIDIA accelerated computing spans the IIT Madras Brain Centre’s technology journey from solution building, rollout, optimization, management, and scaling.
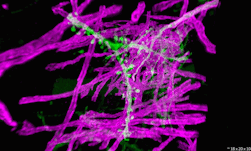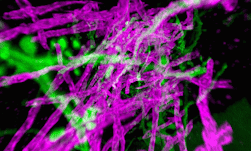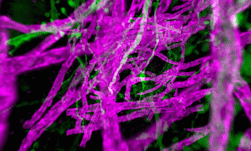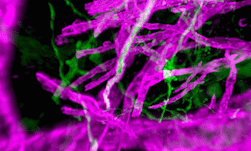
Cellular resolution imaging is readily possible today for smaller brains of insects like flies and mammals like mice and small monkeys. However, the process of acquiring, converting, processing, analyzing, and interpreting, turns into an even more arduous, skill– and time-intensive activity for whole human brains, which are orders of magnitude larger and more complex.
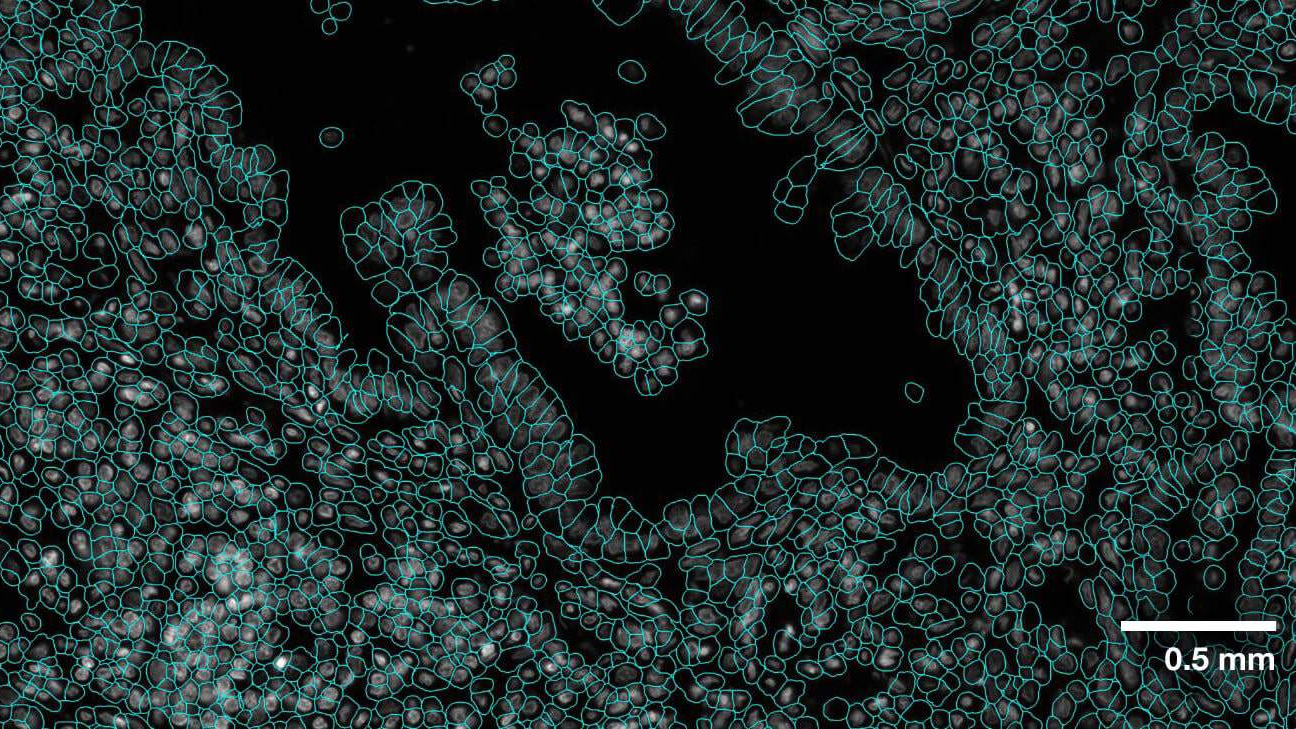
The key big data characteristics of IITM Brain Centre’s imaging pipeline are volume and velocity. At a scanning rate of 250 GB per hour per scanner, operating multiple scanners simultaneously, the center generates 2 TB per hour of high-resolution uncompressed images. All the images must be processed to map every imaged cell. For a computer vision object detection model, the equivalent incidence rate is approximately 10K objects per second.
Handling such large-scale primary neuroanatomy data needs mathematical and computational methods to reveal the complex biological principles governing brain structure, organization, and developmental evolution, at multiple spatio-temporal scales.
This important and challenging scientific pursuit involves the analysis of carefully acquired cellular-resolution brain images for establishing quantitative descriptions of the following:
- Spatial layout
- Cellular composition
- Neuronal pathways
- Compartmental organization
- Whole-brain architecture
It extends to studying inter-brain similarities and relationships at all these levels.
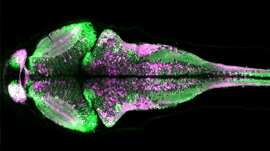
The new Brain Centre of IIT Madras has taken up this challenge and is powering a large-scale, multi-disciplinary effort to map more than 100 human brains at a cellular level. Using their proprietary technology platform, the center is imaging post-mortem human brains of different types and ages.
Their goal is to create an unprecedented cell resolution, uniformly studied, digital collection of multiple types of human brains, which will be queryable from cell level to whole brain level. This requires the capability to enumerate 100B neurons per brain, over 100 brains, and the connectivity across different brain regions.
Meeting the computational demands of cellular resolution brain imaging
The center has developed a world-class computing platform to store, address, access, process, and visualize such high-resolution digital human brain data at the scale of 100+ petabytes, through nothing more than a web browser interface.
This endeavor can be related to the mapping of multiple whole planets to uniformly study patterns, trends, and differences through high-resolution, cross-sectional imaging data through the planet’s volume. Satellite images of the earth’s surface would reach terabytes, a manageable size in today’s computers and web browsers. These geospatial rendering techniques power the likes of Google Maps and others.
However, volumetric imaging of whole brains at cellular resolution yields petabytes of digital data per brain, a challenge to visualize, process, analyze, and query over a web interface.
The computational challenges behind the scenes are equally tremendous:
- A human-in-the-loop large image data pipeline
- Automation right from indexing the data from multiple concurrent imaging systems
- Transporting the images to a central uniform parallel file storage cluster
- Encoding in a format convenient for random access at an arbitrary scale
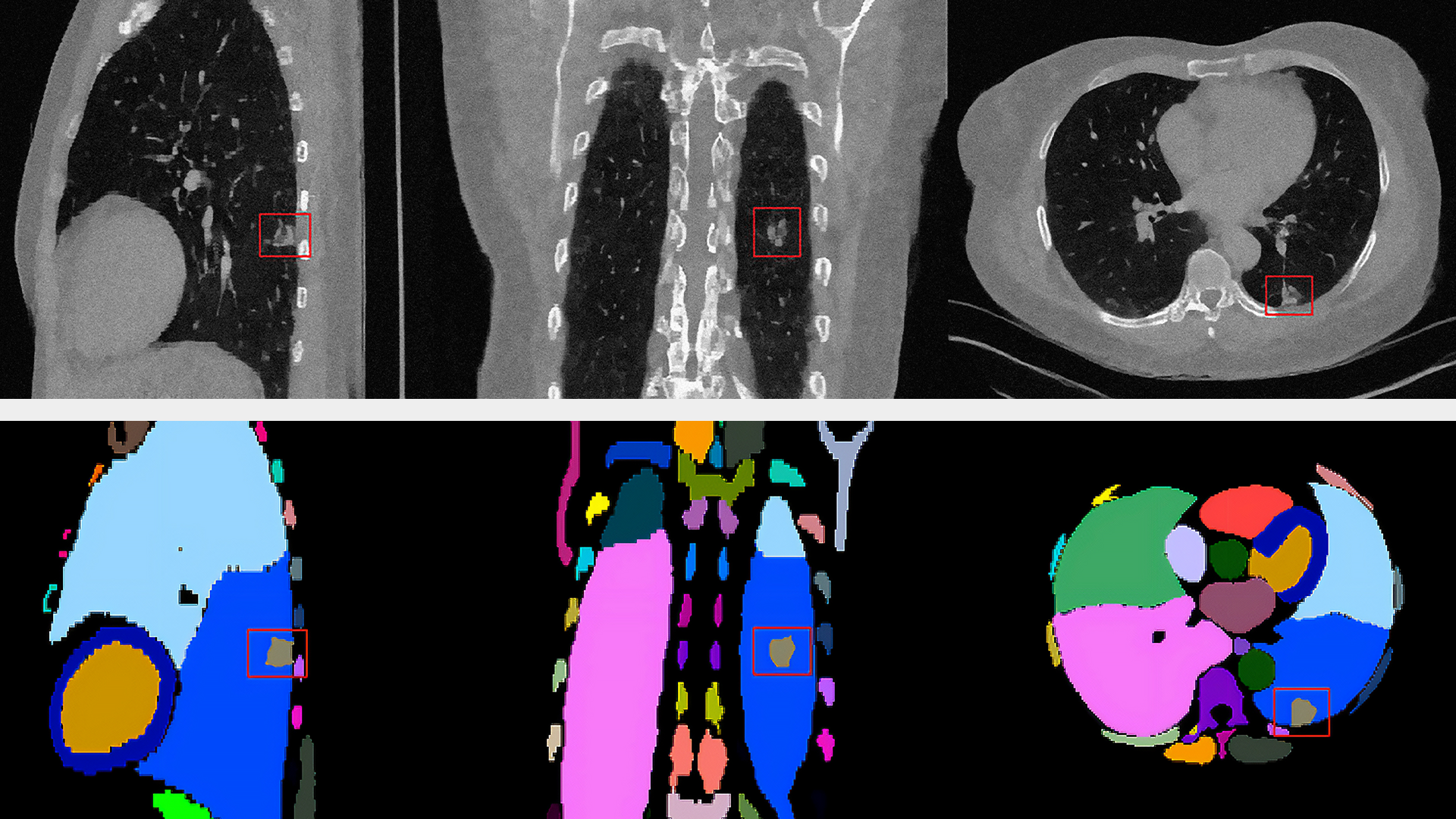
- Machine learning models for multiple automated tasks like the detection of tissue outlines
- Quality control for imaging
- Deep learning tasks for large image normalization
- Cellular-level object detection, classification, and region delineation
- Advanced mathematical models for the geometric alignment of images across modalities and resolutions, followed by computational geometry for deriving quantitative informatics
- Formatting into a high-volume rapid retriable metadata and informatics data store that can transform the prohibitive to the possible: performing single-cell to whole-brain queries on demand and reverting with timely answers for a web-based interaction
To contemplate the scale of the challenge, look at just one of these tasks, a seemingly well-posed task of object detection. This is a task for which modern deep learning convolutional neural networks are known to perform quite well, achieving or in some cases exceeding human-level abilities in identifying and labeling objects.
However, these models are trained to work on megapixel images, containing a few tens of objects. A single-cell resolution image is multi-gigapixel and can contain millions of distinct objects. Processing such large data requires specialized computational workhorses.
This is why the Brain Centre has turned to NVIDIA, a leader in GPU-based HPC offerings, and has operationalized a cluster of DGX A100 systems to do the complete processing of 10 to 20 brains. As the center scales to 100 brains and more, they look to a DGX SuperPOD to provide scalable performance, with industry-leading computing, storage, networking, and infrastructure management.
With eight NVIDIA A100 Tensor Core GPUs per DGX node, the same data that requires a minimum of 1 hour to detect cells has been reduced to less than 10 minutes on an NVIDIA DGX. This enables whole-brain analysis in a month’s time frame, and scaling to 100 brains becomes practical.
“I am delighted to see IITM Brain Centre collaborating with NVIDIA in tackling this challenge of analyzing the very large and complex cellular-level human brain data we are generating,” said Kris Gopalakrishan, a distinguished alumnus of IIT Madras and co-founder of Infosys. He played a pivotal role in setting up and supporting the IITM Brain Centre. “By working with an industry leader like NVIDIA, we look forward to creating breakthroughs in this area leading to global impact.”
Solving the computation challenges
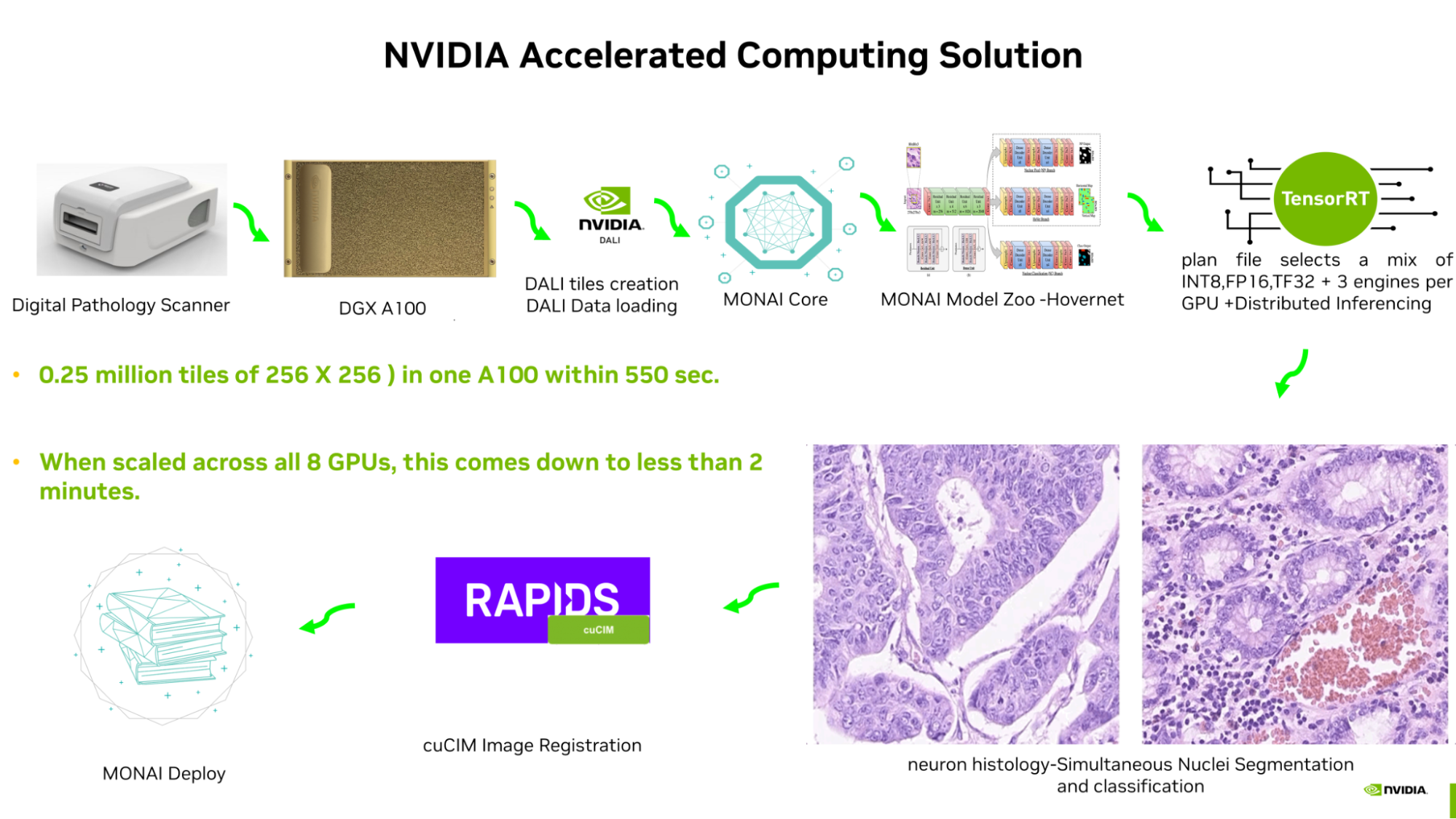
A gigapixel whole slide image making up 0.25 million tiles of 256 X 256 images takes just 420 seconds for inferencing on an A100 GPU. It was possible through end-to-end pipeline optimization using NVIDIA libraries and application frameworks:
- Accelerated tile creation and batching are performed using NVIDIA DALI.
- The MONAI Core model is optimized by TensorRT.
- The TensorRT plan file selects a mix of INT8, FP16, and TF32 at different parts of the network, and produces the engine.
- Three engines are put in one A100 GPU for distributed inferencing.
- The NVIDIA accelerated image processing library cuCIM is used for accelerated image registration.
- NVIDIA IndeX is used for multi-GPU volume visualization for various zoom levels. Soon, MONAI Label’s AI-assisted annotation, along with NVIDIA federated learning SDK-Flare, will be used to further refine various other MONAI core models and the pipeline will be deployed using MONAI Deploy.
“The NVIDIA technology stack empowers the pioneers at the IIT Madras Brain Centre to effectively address the computational needs for high-resolution brain imaging at the cellular level, thereby propelling forward neuroscience research on a national and global scale,” said Vishal Dhupar, managing director of South Asia at NVIDIA.
MONAI and TensorRT are available with NVIDIA AI Enterprise, which is included with the NVIDIA DGX infrastructure.
The compute capability of NVIDIA DGX systems, with dual 64-core CPUs and eight NVIDIA A100 GPUs with 640 GB of GPU memory, along with 2 TB of RAM and 30 TB of flash storage, represent the highest class of server compute available in a single 4U chassis.
Further, the DGX is scalable. NVIDIA offers an ecosystem of software and networking to interconnect multiple DGX systems and match the scale and performance requirements of IITM’s Brain Centre for data processing of pipelined batch jobs, as well as on-demand burst computations.
The effective processing rate of a single NVIDIA A100 GPU for CNN inference as benchmarked on the Brain Centre data is 60 GB per hour (data in uint8, inference in FP16 precision), or 2.4 TB per hour over five DGX servers (40 A100 GPUs), which matches the current imaging rate. This makes pipelining of imaging and computation bottleneck-free. Owing to the scalability of the DGX compute nodes, any surge in data inflow rate can also be matched through scale-out growth.
The A100 GPU is mostly targeted for deep learning training for large datasets and large models that might not fit in smaller GPU vRAM. In the context of the IITM Brain Centre, the A100 GPUs within the DGX systems are used in CNN inference, in a multi-engine per A100 GPU fashion, with the data being mapped out across multi-GPU and multi-node, for scaling from 1–40x across five DGX servers.
This enables the handling of variable image sizes, corresponding to the physical size variability in human brains across ages from fetal to adult (1–32x scale variation). Also, the CPU compute capabilities and the storage type of the DGX A100 systems are well-used in the Brain Centre’s compute pipeline for workloads that are CPU-intensive or data access or movement-intensive, as well as for remote visualization.
The NVIDIA technology stack offers tools and optimized libraries for every step in the process, in a uniform format as container runtimes, facilitating the adoption with minimal effort and ensuring best practices and automated operation.
Transforming the landscape of deep learning in medical imaging
Deep learning technology used to focus on engineering the best methods or tuning at training time for incremental boosts in performance. It has now shifted to inference with proven foundational models across domains like computer vision (object detection, semantic and panoptic segmentation, DL-based image registration) and natural language. The results are realizing applications previously held as non-amenable for computational automation.
The emphasis now is on implementing software guardrails around new applications powered by deep learning inference. Integrated hardware systems and software stacks are not just a convenience in this new direction but are a vehicle of scale and simplification. The NVIDIA technology stack is a way to leapfrog solution building, deployment, and scaling.
A detailed map of the earth is today accessible for everyone and has emerged as a platform enabling new applications, and businesses. It now guides and shapes globally how we move about. The efforts of the IIT Madras Brain Centre are targeted to produce a similar, transformative platform that will yield new outcomes in brain science, shape and guide brain surgery and therapy, and expand our understanding of medicine’s last frontier, the human brain.
For more information, see the /HTIC-Medical-Imaging GitHub repo.