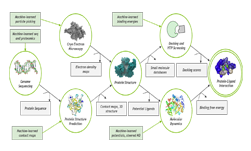
Computational molecular design involves compute-intense calculations that require exceptional processing power.
Whether working in pharmaceuticals, biotechnology, agrochemicals, or the fragrance industry, researchers are oftentimes dealing with datasets that encompass millions to billions of compounds.
Until recently, this required that companies invest in expensive, specialized high-performance computing (HPC) capabilities to undertake large-scale molecular design programs. For many teams, this meant that their research was frequently limited by hardware and budget constraints.
With the advent of GPUs, fast, accurate, and cost-sensitive calculations are now more accessible than ever.
OpenEye Scientific, an industry leader in scientific computing and molecular design, has updated several of its products to leverage the power of GPUs. As a result, research groups of any size are now better positioned to perform large-scale molecular calculations and simulations, to quickly analyze exponentially larger datasets, and to increase the speed of calculations while also reducing associated costs.
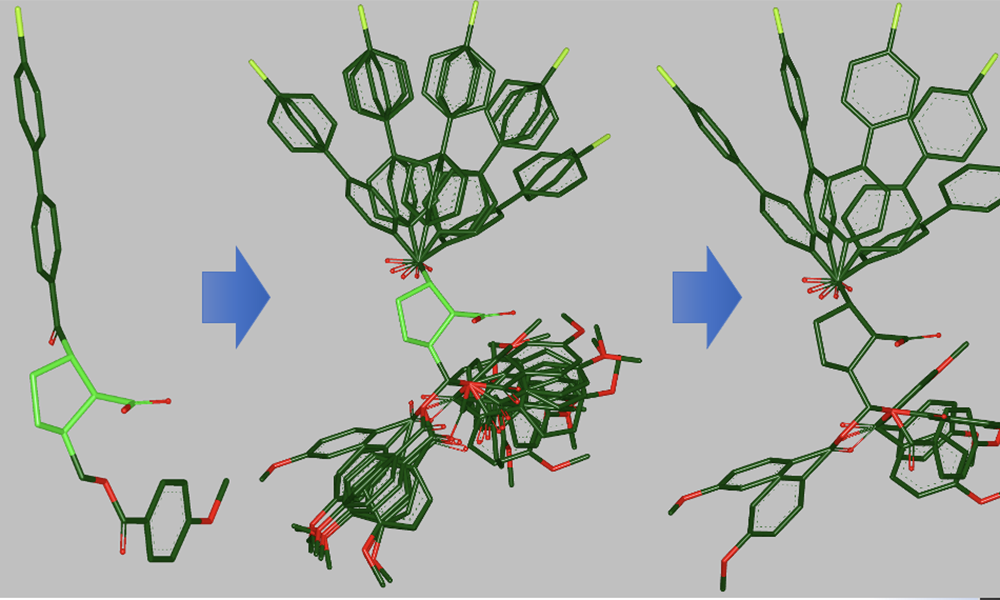
At the heart of many OpenEye products is OMEGA. The tool uses 1D or 2D molecular data files to generate high-quality and bioactive 3D conformers, which are different shapes of the 3-dimensional molecular structure.
Researchers then use these 3D conformers to perform various molecular modeling or drug discovery tasks with other OpenEye software products, such as:
- Shape and chemical feature similarity scoring (ROCS® and FastROCS™) Electrostatic scoring (EON)
- Fragment replacement (BROOD)
- Docking and posing (OEDocking)
- Force field optimization (SZYBKI)
- Ligand thermodynamics (FreeForm)
OpenEye recently has updated OMEGA to work with NVIDIA market-leading GPU processors. Running OMEGA with GPUs results in 30X faster conformer generation compared to traditional CPU processing time. It also reduces costs compared to running such large-scale processing using traditional CPUs.
Benchmarking Results
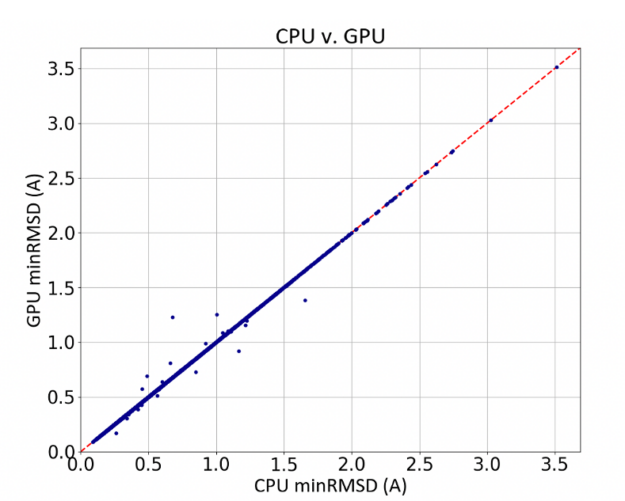
Benchmarking studies show that running GPU-OMEGA, compared to traditional CPU, results in significantly faster conformer generation, with no accuracy loss.
In one comparison, GPU-OMEGA performs at the same high level of accuracy as OpenEye’s original OMEGA software, as shown below using a filtered subset of the Platinum dataset [Friedrich-2017] [Hawkins-2020].
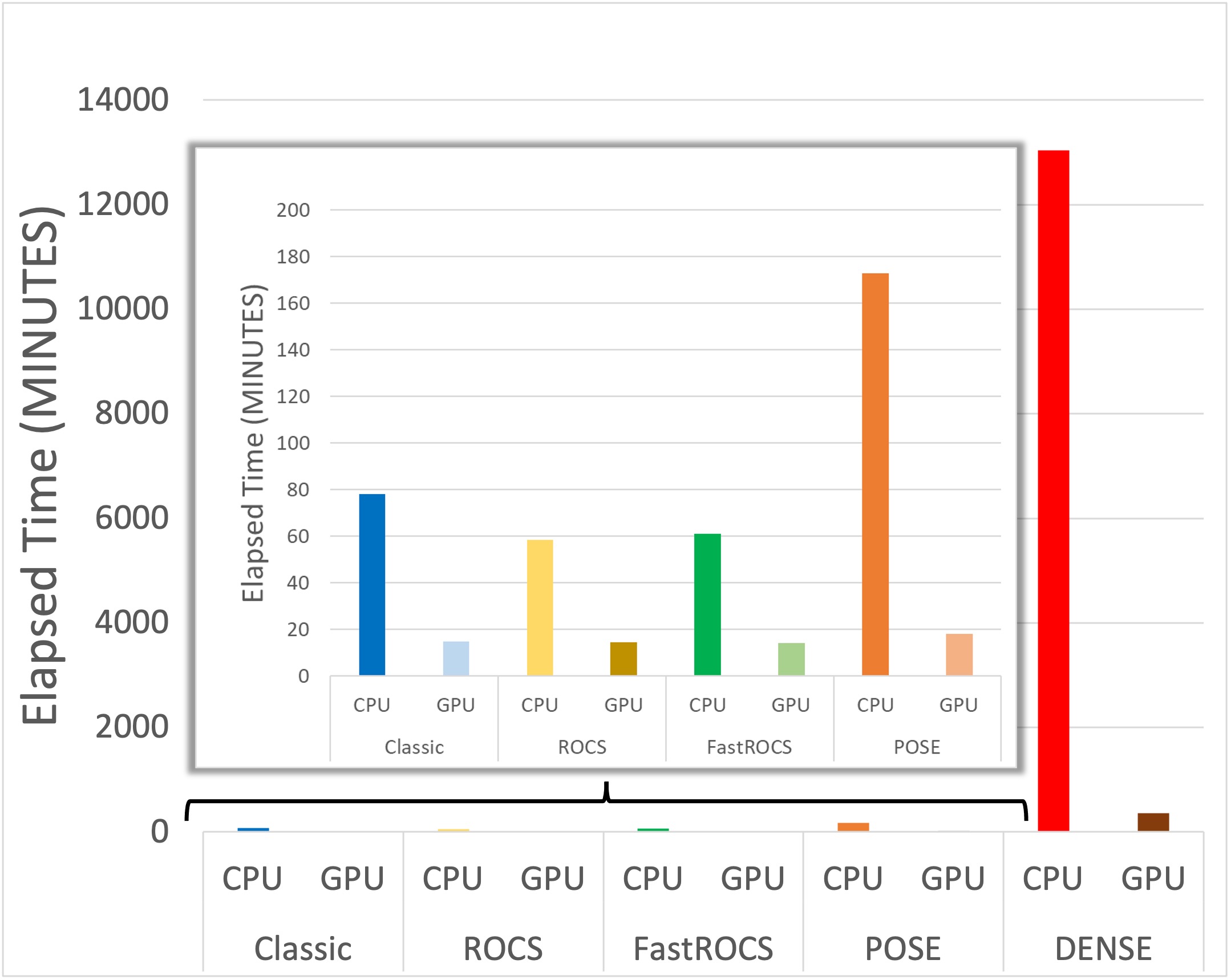
GPU-OMEGA was also benchmarked against the original CPU version of OMEGA with a single NVIDIA A100 GPU on all the available sampling modes (Classic, DENSE, POSE, ROCS, and FastROCS) using a subset of approximately 13,000 molecules from the GSK TCAMs dataset [Gamo-2010]. The most impressive speed-up is seen with the dense sampling mode, with a reduction in processing time from nearly 220 hours to just more than 6 hours.
Learn More about OMEGA
Learn more about OpenEye Scientific’s OMEGA molecular conformer generator.