NVIDIA Parabricks v4.3 was released at NVIDIA GTC 2024, introducing new tooling and workflows that bring acceleration and the latest AI techniques to multiple omics data types. In addition to analyzing DNA and RNA, you can now also analyze methylation, single-cell, and spatial omics workloads at high speed and high accuracy with the power of GPUs and generative AI.
Parabricks v4.3 also drives analysis time down more than ever before, with further optimizations to germline tools, and support for the latest GPU architectures.
What’s new
- A brand-new workflow for single-cell and spatial omics data that incorporates software from across NVIDIA (including imaging, generative AI, and accelerated data science) to enable rapid and high-accuracy analysis.
- An accelerated version of BWA-Meth, as a new tool in Parabricks to accelerate bisulfite sequencing alignment for DNA methylation data.
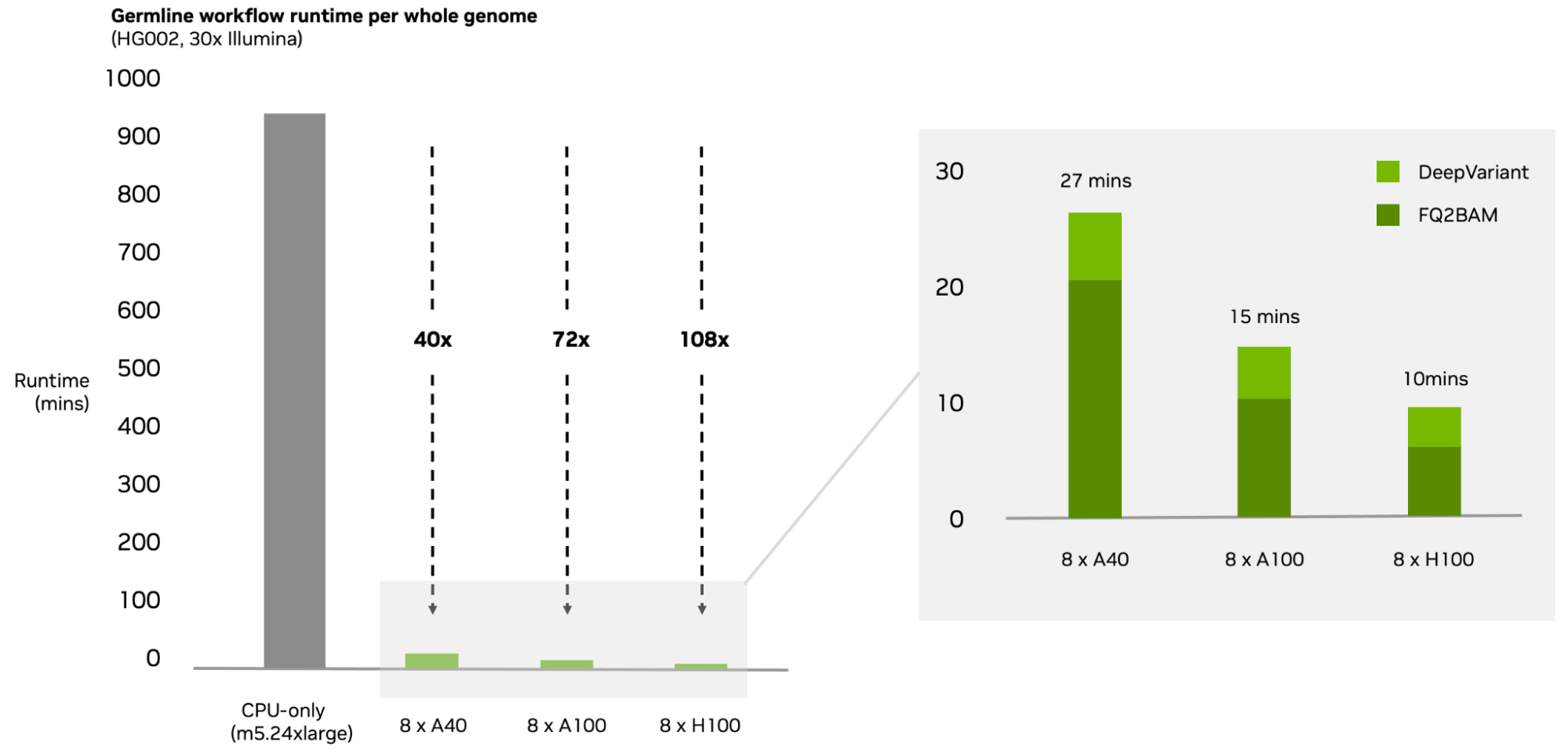
- Further optimized germline analysis, using the NVIDIA H100 Dynamic Programming Core to bring germline analysis down to under 10 minutes, now the fastest application of gold-standard tools (BWA-MEM and GATK) on the market
- Support for the latest NVIDIA data center GPUs, including NVIDIA Grace Hopper
- An upgraded version of DeepVariant to v1.6, to support NovaSeqX data
Single-cell and spatial omics
There are techniques that enable researchers to understand omics at a cellular level, such as single-cell sequencing, and to put that data into the context of a tissue with spatially resolved approaches. These techniques are becoming increasingly popular as a way to model the biological systems underpinning cells and tissues.
This poses several new challenges for the industry.
First, the scale of the data being produced in single-cell and spatial experiments is skyrocketing. Single-cell atlas projects are reaching the order of millions of cells and the latest generations of spatial omics instruments are producing on the order of petabytes of imaging data.
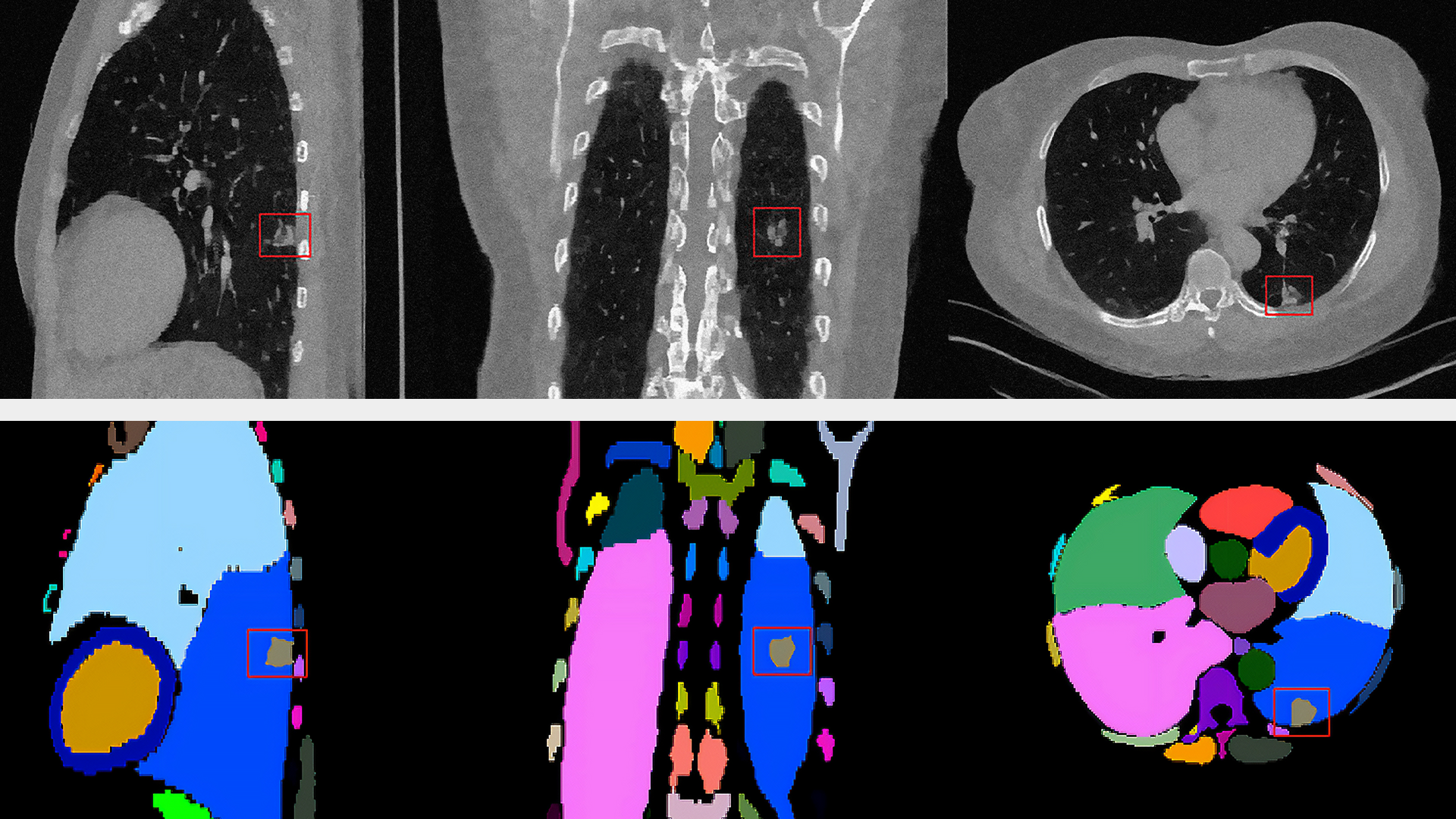
Second, the analysis of this requires much more automation. Image-based spatial approaches require cells to be segmented and expression values quantified in up to hundreds of thousands of cells. Segmentation is a non-trivial task with many approaches, including human-in-the-loop, to guide algorithms when faced with a new dataset or cell type.
Finally, the data produced provides an entirely new potential for insights, with not just expression data available, but also morphology of the imaged cells providing potentially useful information to researchers.
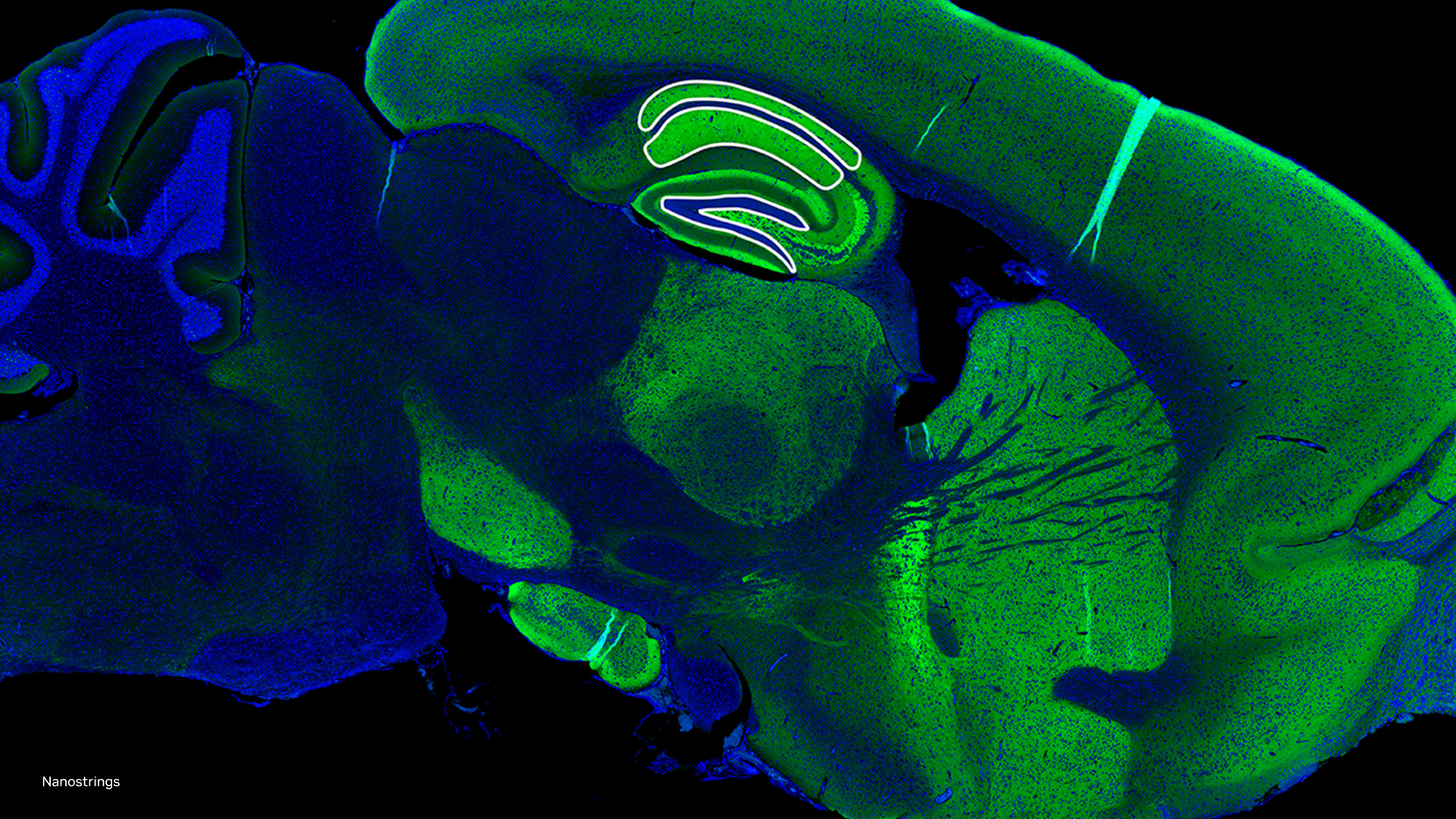
Spatial omics providers like Nanostring, are already using NVIDIA GPUs and accelerating the compute onboard their CosMx SMI device to address these challenges.
“Spatial-omics technology (CosMx Spatial Molecular Imaging) can now image the entire transcriptome inside cells and tissues, generating data at a density and scale that has never before been accomplished (~ 150 TB per cm^2). This data will play a critical role in transforming our understanding of both health and disease, radically accelerating drug development and spatial diagnostics,” said Joseph Beechem, Ph.D., chief scientific officer and senior vice president of research and development, NanoString.
“In reality, generative AI will be required to explore the true information content of these images. We’re thrilled to continue deepening our partnership with NVIDIA at all levels of the data-to-information pipeline. We invite all of the AI/ML community to join us in this Life Sciences Spatial Biology revolution.”
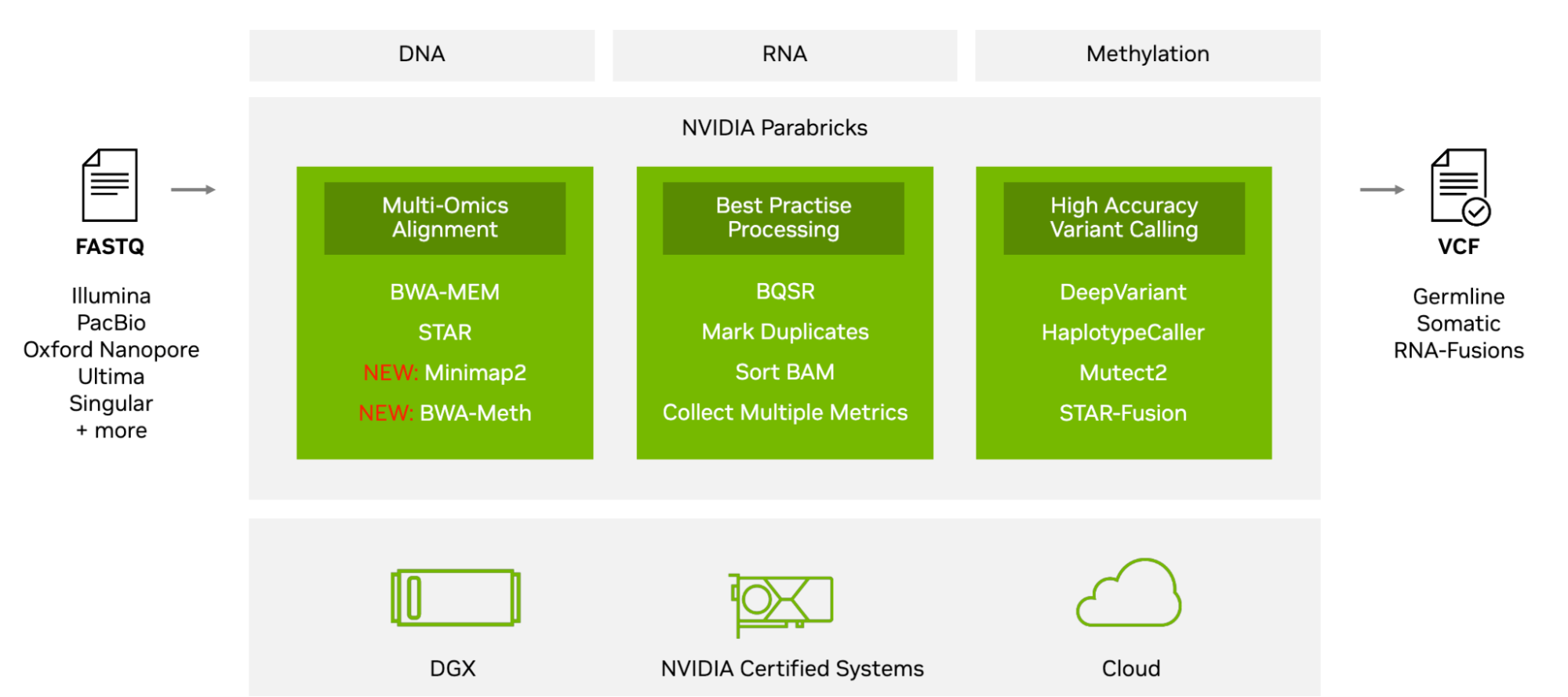
To further build on this, NVIDIA Parabricks now includes a single-cell and spatial omics reference workflow (Figure 1), to accelerate computational bottlenecks, provide a greater level of accuracy, and enable novel methods of analysis.
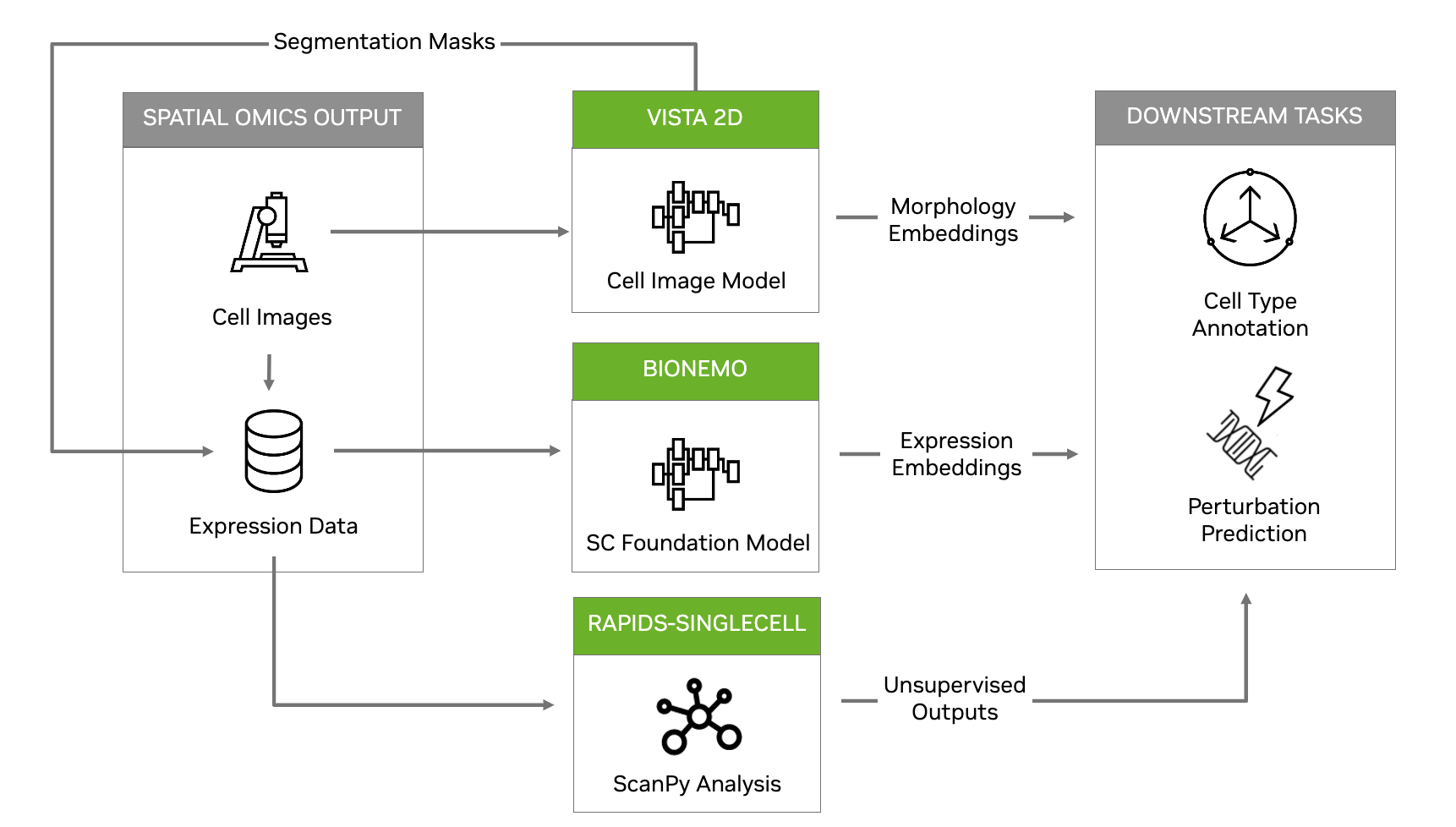
For addressing bottlenecks in the analysis of single-cell and spatial expression outputs, NVIDIA recently collaborated with Severin Dicks from Charité Berlin to bring NVIDIA RAPIDS to the scverse ecosystem. The RAPIDS-singlecell library primarily designed as an accelerated drop-in replacement for Scanpy, enabling accelerations of up to 850x on a single A100 GPU in some cases.
NVIDIA is now bringing generative AI to single-cell and spatial omics analysis, with a new cell imaging foundation model capable of high-accuracy segmentation (highly important for correctly attributing expression to the correct cell, affecting the accuracy of all downstream tasks) and also uniquely capable of generating embeddings representing the cell morphology.
The NVIDIA BioNeMo Framework is also releasing capabilities for building and training single-cell BERT models, for applying generative capabilities such as perturbation prediction to single-cell expression data.
All are now combined in a reference workflow provided and maintained on the public /clara-parabricks-workflows GitHub repo.
Parabricks v4.3 tools and benchmarks
In addition to developments in single-cell and spatial, we released NVIDIA Parabricks version 4.3, furthering the Parabricks mission to accelerate alignment and variant calling across all sequencers and omics.
This release goes even further to break the bottleneck of sequencing analysis, optimizing the following industry-trusted tools:
- BWA-MEM
- GATK BQSR
- MarkDuplicates
- HaplotypeCaller
- DeepVariant
Through development using NVIDIA H100 DPX instructions, this Parabricks release has driven down runtimes of end-to-end analysis to achieve groundbreaking results.
Engineers at Oracle Cloud are already succeeding in running this workflow in record times. “We are delighted by the continued acceleration by NVIDIA for genomic analysis from both a hardware and software perspective,” said Dan Spellman, global AI cloud director for Oracle. “Using the latest version of Parabricks and H100, OCI’s genomics engineering led by Ruzhu Chen has reduced the processing time to now below 10 minutes.”
Further development of tooling in version 4.3 includes runtime improvements to GPU-accelerated Minimap2 for aligning PacBio data and an accelerated version of BWA-Meth for methylation data alignment called fq2bam_meth.
DNA methylation is a key component of the epigenome, playing an important role in regulating gene expression in different tissues. Research into methylation and the epigenome has shown these can be causal of disease and can provide key markers in applications such as cardiovascular disease testing, or liquid biopsies for early detection of cancer from blood.
BWA-Meth is a tool for accurate alignment of bisulfite-converted DNA reads (a process of exposing methylation by converting all non-methylated cytosines to uracils). Acceleration of BWA-Meth in Parabricks can achieve up to 36x acceleration on NVIDIA DGX A100 (eight NVIDIA A100 GPUs) compared to CPU alone (c5.12xlarge, 48 CPU threads), running on whole genome bisulfite sequencing data.
Researchers at Sequanta, a multi-omics research and clinical service provider in China, have already achieved 21x acceleration with Parabricks BWA-Meth over other alignment approaches. They have now been able to perform alignment on methylation samples in just 60 minutes on eight T4 GPUs, as opposed to other approaches taking 21 hours.
Learn more
NVIDIA Parabricks v4.3 offers accelerated multi-omics analysis, addressing challenges in DNA, RNA, methylation, single-cell, and spatial omics data. It provides a reference workflow for single-cell and spatial omics, accelerating analysis and enabling novel methods.
With optimizations and support for industry-trusted tools, Parabricks reduces runtime to under 10 minutes, breaking the bottleneck of sequencing analysis.
Download Parabricks v4.3 containers from NGC and access the reference workflows on GitHub to accelerate your multi-omics analysis and gain deeper insights into biological systems. Get started with Parabricks today and unlock the potential of accelerated genomic analysis.