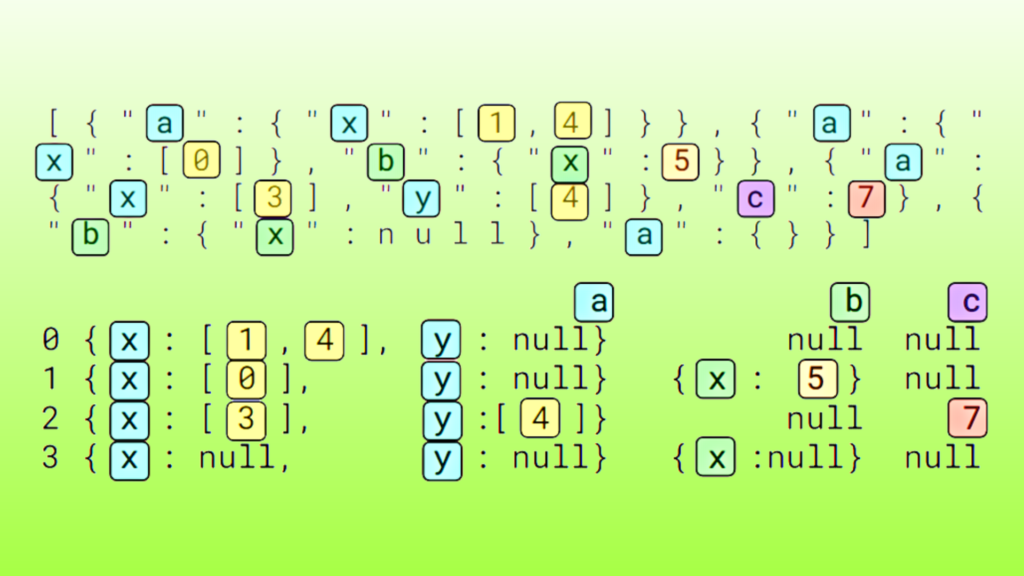
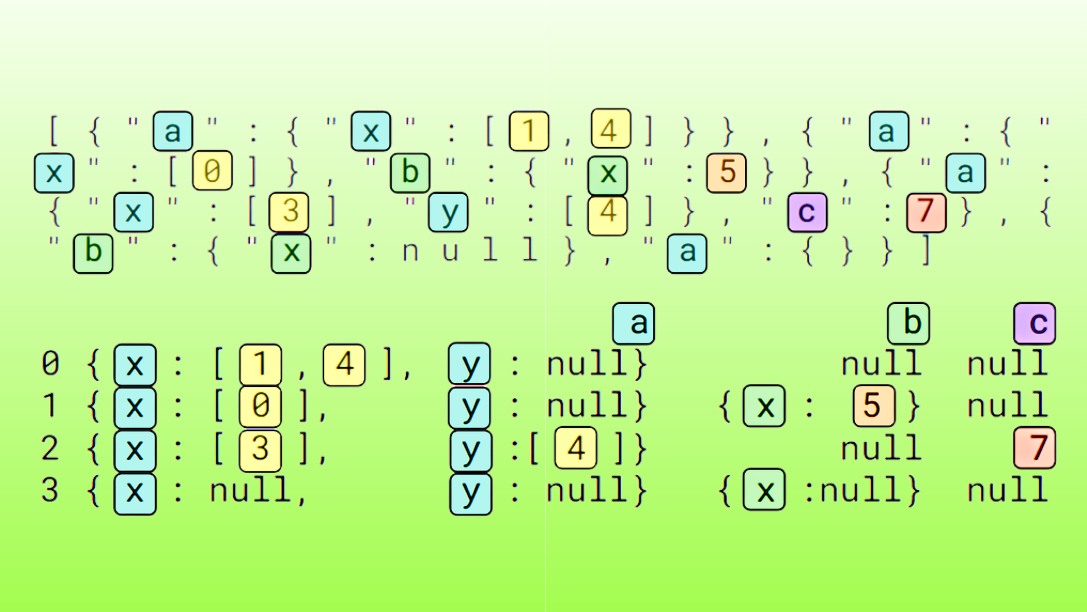
JSON is a widely adopted format for text-based information working interoperably between systems, most commonly in web applications and large language models (LLMs). While the JSON format is human-readable, it is complex to process with data science and data engineering tools.
JSON data often takes the form of newline-delimited JSON Lines (also known as NDJSON) to represent multiple records in a dataset. Reading JSON Lines data into a dataframe is a common first step in data processing.
In this post, we compare the performance and functionality of Python APIs for converting JSON Lines data into a dataframe using the following libraries:
- pandas
- DuckDB
- pyarrow
- RAPIDS cuDF pandas Accelerator Mode
We demonstrate good scaling performance and high data processing throughput with the JSON reader in cudf.pandas, especially for data with a complex schema. We also review the versatile set of JSON reader options in cuDF that improve compatibility with Apache Spark and empower Python users to handle quote normalization, invalid records, mixed types and other JSON anomalies.
JSON parsing versus JSON reading
When it comes to JSON data processing, it’s important to distinguish between parsing and reading.
JSON parsers
JSON parsers, such as simdjson, convert a buffer of character data into a vector of tokens. These tokens represent the logical components of JSON data, including field names, values, array begin/end, and map begin/end. Parsing is a critical first step in extracting information from JSON data, and significant research has been dedicated to reaching high parsing throughput.
To use information from JSON Lines in data processing pipelines, the tokens must often be converted into a Dataframe or columnar format, such as Apache Arrow.
JSON readers
JSON readers, such as pandas.read_json convert input character data into a Dataframe organized by columns and rows. The reader process begins with a parsing step and then detects record boundaries, manages the top-level columns and nested struct or list child columns, handles missing and null fields, infers data types, and more.
JSON readers convert unstructured character data into a structured Dataframe, making JSON data compatible with downstream applications.
JSON Lines reader benchmarking
JSON Lines is a flexible format for representing data. Here are some important properties of JSON data:
- Number of records per file
- Number of top level columns
- Depth of struct or list nesting for each column
- Data types of values
- Distribution of string lengths
- Fraction of missing keys
For this study, we held the record count fixed at 200K and swept the column count from 2 to 200, exploring a range of complex schemas. The four data types in use are as follows:
list<int>andlist<str>with two child elementsstruct<int>andstruct<str>with a single child element
Table 1 shows the first two columns of the first two records for data types, including list<int>, list<str>, struct<int>, and struct<str>.
| Data type | Example records |
|---|---|
list<int> | {"c0":[848377,848377],"c1":[164802,164802],...\n{"c0":[732888,732888],"c1":[817331,817331],... |
list<str> | {"c0":["FJéBCCBJD","FJéBCCBJD"],"c1":["CHJGGGGBé","CHJGGGGBé"],...\n{"c0":["DFéGHFéFD","DFéGHFéFD"],"c1":["FDFJJCJCD","FDFJJCJCD"],... |
struct<int> | {"c0":{"c0":361398},"c1":{"c0":772836},...\n{"c0":{"c0":57414},"c1":{"c0":619350},... |
struct<str> | {"c0":{"c0":"FBJGGCFGF"},"c1":{"c0":"ïâFFéâJéJ"},...\n{"c0":{"c0":"éJFHDHGGC"},"c1":{"c0":"FDâBBCCBJ"},... |
Table 1 shows the first two columns of the first two records for data types, including list<int>, list<str>, struct<int>, and struct<str>.
Performance statistics were collected on the 25.02 branch of cuDF and with the following library versions: pandas 2.2.3, duckdb 1.1.3, and pyarrow 17.0.0. The execution hardware used an NVIDIA H100 Tensor Core 80 GB HBM3 GPU and Intel Xeon Platinum 8480CL CPU with 2TiB of RAM. Timing was collected from the third of three repetitions, to avoid initialization overhead and ensure that the input file data was present in the OS page cache.
In addition to the zero code change cudf.pandas, we also collected performance data from pylibcudf, a Python API for the libcudf CUDA C++ computation core. The runs with pylibcudf used a CUDA async memory resource through RAPIDS Memory Manager (RMM). Throughput values were computed using the JSONL input file size and the reader runtime of the third repetition.
Here are some examples from several Python libraries for invoking the JSON Lines reader:
# pandas and cudf.pandas
import pandas as pd
df = pd.read_json(file_path, lines=True)
# DuckDB
import duckdb
df = duckdb.read_json(file_path, format='newline_delimited')
# pyarrow
import pyarrow.json as paj
table = paj.read_json(file_path)
# pylibcudf
import pylibcudf as plc
s = plc.io.types.SourceInfo([file_path])
opt = plc.io.json.JsonReaderOptions.builder(s).lines(True).build()
df = plc.io.json.read_json(opt)
JSON Lines reader performance
Overall, we found a wide range of performance characteristics for the JSON readers available in Python, with overall runtimes varying from 1.5 seconds to almost 5 minutes.
Table 2 shows the sum of the timing data from seven JSON reader configurations when processing 28 input files with a total file size of 8.2 GB:
- Using cudf.pandas for JSON reading shows about 133x speedup over pandas with the default engine and 60x speedup over pandas with the pyarrow engine.
- DuckDB and pyarrow show good performance as well, with about 60 seconds total time for DuckDB, and 6.9 seconds for pyarrow with block size tuning.
- The fastest time comes from pylibcudf at 1.5 seconds, showing about 4.6x speedup over pyarrow with
block_sizetuning.
| Reader label | Benchmark runtime (sec) | Comment |
| cudf.pandas | 2.1 | Using -m cudf.pandas from the command line |
| pylibcudf | 1.5 | |
| pandas | 281 | |
| pandas-pa | 130 | Using the pyarrow engine |
| DuckDB | 62.9 | |
| pyarrow | 15.2 | |
| pyarrow-20MB | 6.9 | Using a 20 MB block_size value |
Table 2 includes the input columns counts 2, 5, 10, 20, 50, 100, and 200, and the data types list<int>, list<str>, struct<int>, and struct<str>.
Zooming into the data by data type and column count, we found that JSON reader performance varies over a wide range based on the input data details and the data processing library, from 40 MB/s to 3 GB/s for CPU-based libraries and from 2–6 GB/s for the GPU-based cuDF.
Figure 1 shows the data processing throughput based on input size for 200K rows and 2–200 columns, with input data sizes varying from about 10 MB to 1.5 GB.

In Figure 1, each subplot corresponds to the data type of the input columns. File size annotations align to the x-axis.
For cudf.pandas read_json, we observed 2–5 GB/s throughput that increased with larger column count and input data size. We also found that the column data type does not significantly affect throughput. The pylibcudf library shows about 1–2 GB/s higher throughput than cuDF-python, due to lower Python and pandas semantic overhead.
For pandas read_json, we measured about 40–50 MB/s throughput for the default UltraJSON engine (labeled as “pandas-uj”). Using the pyarrow engine (engine="pyarrow") provided a boost up to 70–100 MB/s due to faster parsing (pandas-pa). The pandas JSON reader performance appears to be limited by the need to create Python list and dictionary objects for each element in the table.
For DuckDB read_json, we found about 0.5–1 GB/s throughput for list<str> and struct<str> processing with lower values <0.2 GB/s for list<int> and struct<int>. Data processing throughput remained steady over the range of column counts.
For pyarrow read_json, we measured data processing throughputs up to 2–3 GB/s for 5-20 columns, and lower throughput values as column count increased to 50 and above. We found data type to have a smaller impact on reader performance than column count and input data size. For column counts of 200 and a record size of ~5 KB per row, throughput dropped to about 0.6 GB/s.
Raising the pyarrow block_size reader option to 20 MB (pyarrow-20MB) led to increased throughput for column counts 100 or more, while also degrading throughput for column counts 50 or fewer.
Overall, DuckDB primarily showed throughput variability due to data types, whereas cuDF and pyarrow primarily showed throughput variability due to column count and input data size. The GPU-based cudf.pandas and pylibcudf showed the highest data processing throughput for complex list and struct schema, especially for input data sizes >50 MB.
JSON Lines reader options
Given the text-based nature of the JSON format, JSON data often includes anomalies that result in invalid JSON records or don’t map well to a dataframe. Some of these JSON anomalies include single-quoted fields, cropped or corrupted records, and mixed struct or list types. When these patterns occur in your data, they can break the JSON reader step in your pipeline.
Here are some examples of these JSON anomalies:
# 'Single quotes'
# field name "a" uses single quotes instead of double quotes
s = '{"a":0}\n{\'a\':0}\n{"a":0}\n'
# ‘Invalid records'
# the second record is invalid
s = '{"a":0}\n{"a"\n{"a":0}\n'
# 'Mixed types'
# column "a" switches between list and map
s = '{"a":[0]}\n{"a":[0]}\n{"a":{"b":0}}\n'
To unlock advanced JSON reader options in cuDF, we recommend incorporating cuDF-Python (import cudf) and pylibcudf into your workflow. If single-quoted field names or string values appear in your data, cuDF provides a reader option to normalize single quotes into double quotes. cuDF supports this feature to provide compatibility with the allowSingleQuotes option that is enabled by default in Apache Spark.
If invalid records appear in your data, cuDF and DuckDB both provide error recovery options to replace these records with null. When error handling is enabled, if a record generates a parsing error, all of the columns for the corresponding row are marked as null.
If mixed list and struct values are associated with the same field name in your data, cuDF provides a dtype schema override option to coerce the datatype to string. DuckDB uses a similar approach by inferring a JSON data type.
For mixed types, the pandas library has perhaps the most faithful approach, using Python list and dictionary objects to represent the input data.
Here is an example in cuDF-Python and pylibcudf that shows the reader options, including a dtype schema override for column name “a”. For more information, see cudf.read_json and pylibcudf.io.json.read_json.
For pylibcudf, the JsonReaderOptions object can be configured either before or after the build function.
# cuDF-python
import cudf
df = cudf.read_json(
file_path,
dtype={"a":str},
on_bad_lines='recover',
lines=True,
normalize_single_quotes=True
)
# pylibcudf
import pylibcudf as plc
s = plc.io.types.SourceInfo([file_path])
opt = (
plc.io.json.JsonReaderOptions.builder(s)
.lines(True)
.dtypes([("a",plc.types.DataType(plc.types.TypeId.STRING), [])])
.recovery_mode(plc.io.types.JSONRecoveryMode.RECOVER_WITH_NULL)
.normalize_single_quotes(True)
.build()
)
df = plc.io.json.read_json(opt)
Table 3 summarizes the behavior of several JSON readers with Python APIs for a few common JSON anomalies. Crosses denote that the reader function raised an exception, and checkmarks denote that the library successfully returned a Dataframe. These results may change in future versions of the libraries.
| Single quotes | Invalid records | Mixed types | |
| cuDF-Python, pylibcudf | ✔️ Normalize to double quotes | ✔️ Set to null | ✔️ Represent as a string |
| pandas | ❌ Exception | ❌ Exception | ✔️ Represent as a Python object |
pandas (engine="pyarrow“) | ❌ Exception | ❌ Exception | ❌ Exception |
| DuckDB | ❌ Exception | ✔️ Set to null | ✔️ Represent as a JSON string-like type |
| pyarrow | ❌ Exception | ❌ Exception | ❌ Exception |
cuDF supports several additional JSON reader options that are critical for compatibility with Apache Spark conventions, and now are available to Python users as well. Some of these options include:
- Validation rules for numbers and strings
- Custom record delimiters
- Column pruning by the schema provided in dtype
- Customization of NaN values
For more information, see the libcudf C++ API documentation on json_reader_options.
For more information about multi-source reading for efficiently processing many smaller JSON Lines files, or byte-range support for breaking up large JSON Lines files, see GPU-Accelerated JSON Data Processing with RAPIDS.
Summary
RAPIDS cuDF provides powerful, flexible, and accelerated tools for working with JSON data in Python.
GPU-accelerated JSON data processing is also available in RAPIDS Accelerator For Apache Spark, starting in the 24.12 release. For information, see Accelerating JSON Processing on Apache Spark with GPUs.
For more information, see the following resources:
- cuDF documentation
- /rapidsai/cudf GitHub repo
- RAPIDS Docker containers (available for releases and nightly builds)
- Accelerate Data Science Workflows with Zero Code Changes DLI course
- Mastering the cudf.pandas Profiler for GPU Acceleration