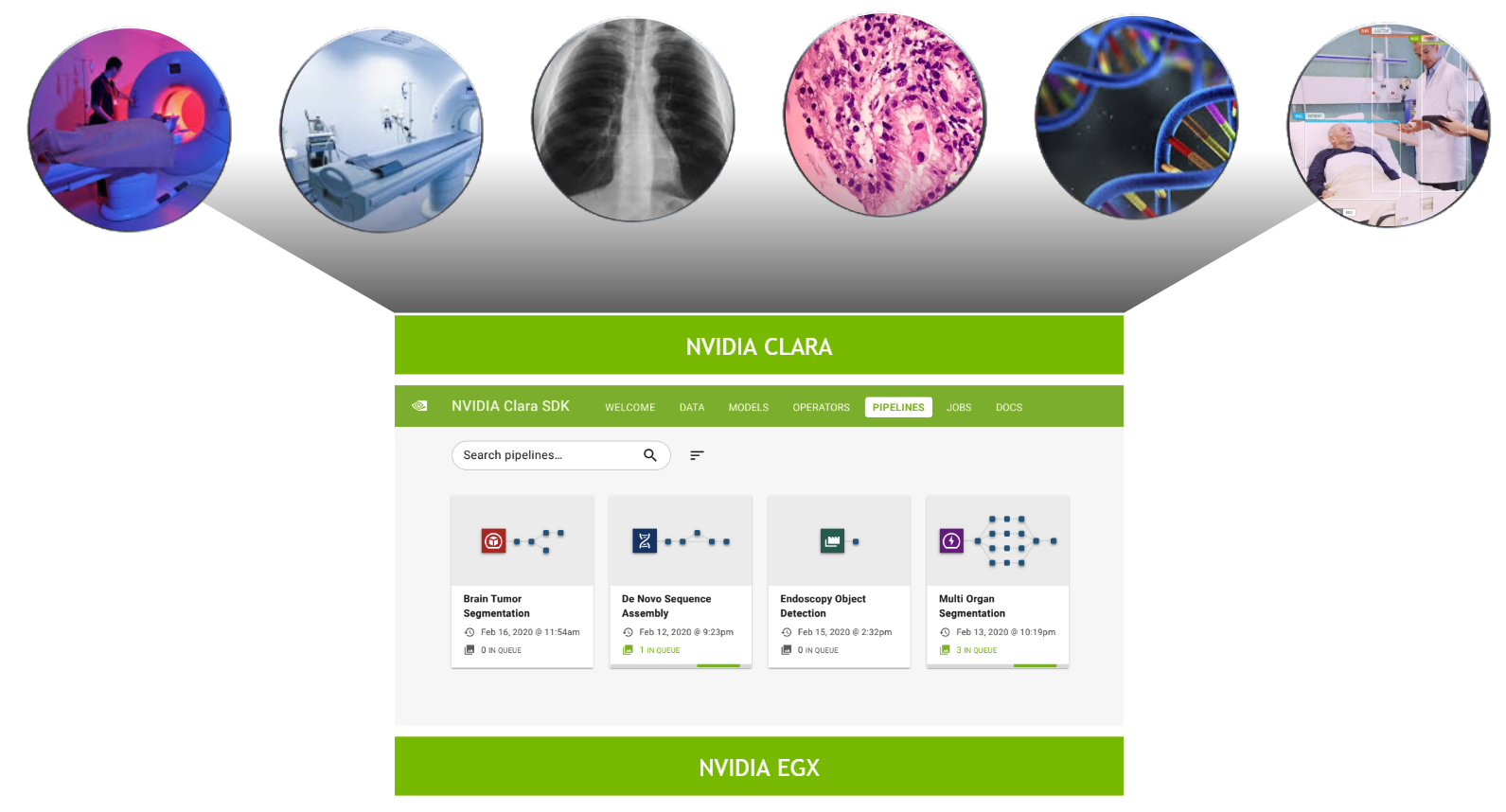
The adoption of AI in hospitals is accelerating rapidly. There are many reasons for this. With Moore’s law broken and computational capability ever increasing, models that save lives and make us more efficient and effective are becoming the norm. Within the next five years, we will see the rise of the “smart hospital,” augmented by workflows incorporating thousands of AI models.
These smart hospitals adopting AI applications face big challenges in IT and infrastructure. Healthcare demands specific restrictions in how data is transmitted, and respecting patient data privacy is paramount. Flexible compute capability, with “write once, run anywhere” capability makes it possible to deploy state-of-the-art applications at the edge in hospitals. Each application demands different compute capabilities for HPC, AI, and visualization.
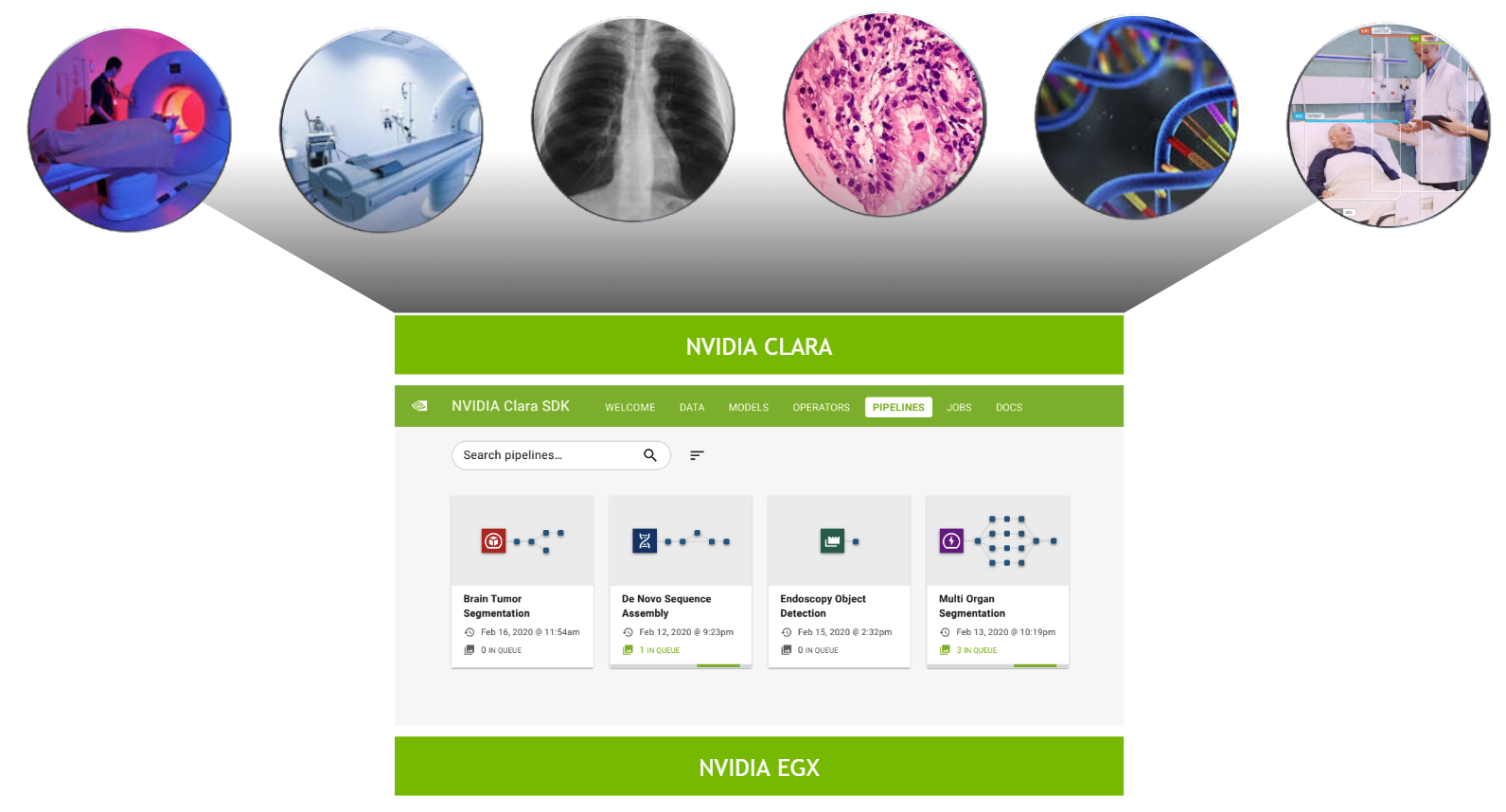
The NVIDIA Clara Deploy SDK answers this call by providing a reference framework for the deployment of multi-AI, multi-modality workflows in smart hospitals: one architecture orchestrating and scaling imaging, genomics, and video processing workloads.
The most pressing problem for deploying AI models is architecting an inference platform that can handle the rapidly changing AI ecosystem, including the increasing number of requests for processing, massive size of healthcare datasets, and diversity of the processing pipelines themselves that use a heterogeneous computing environment.
During GTC Digital 2020, we released an update to the Clara Deploy SDK that included platform features and reference applications that enable developers and data scientists with a unified foundation for delivering intelligent workloads and realizing the vision of the smart hospital.
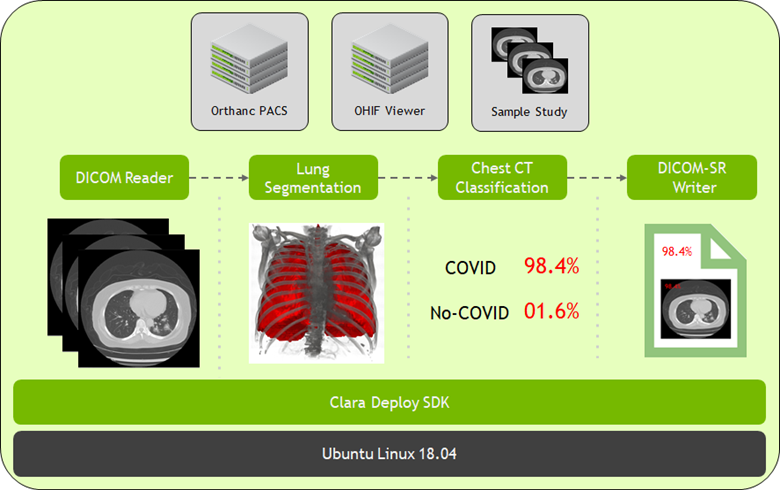
At SIIM 2020, we have now made available the latest version for Clara Deploy SDK, which includes new reference pipelines for COVID-19 inference from CT datasets, a suite of tools for digital pathology (including a pipeline, operators, and visualization), and additional tools to accelerate developers (including shared memory for multi-AI pipelines and features like easier DICOM configuration over REST APIs). Figure 2 shows the Clara Deploy SDK technology stack.
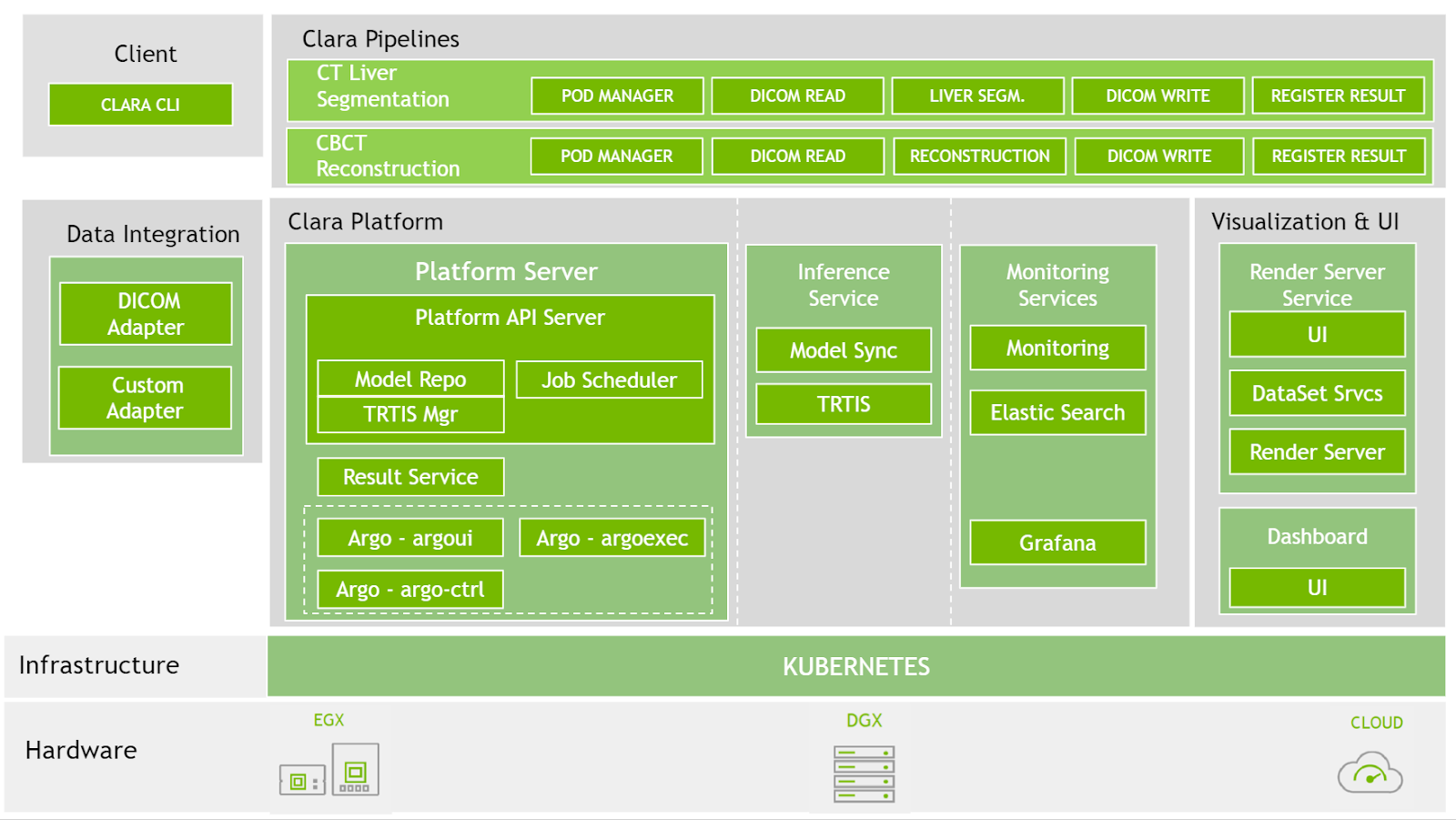
Platform features
The Clara Deploy SDK capabilities include the following:
- Strongly typed operator interface
- Scheduler
- Model repository
- CLI load generator
- EGX support
- Fast I/O integrated with the NVIDIA Clara platform driver
- Configuration of the DICOM adapter
- Distribution of Clara Deploy in NVIDIA NGC
Strongly typed operator interface
In a Clara Deploy SDK pipeline, operators are used to perform each operation. To simplify the development effort and eliminate the guesswork in interfacing with one another, these operators are strongly typed. You can be confident that what you build hangs seamlessly together.

The Clara Deploy SDK supports pipeline composition using operators that conform to a signature, or well-defined interface. This enables the following functionality:
- Pre-runtime validation of pipelines
- Compatibility of concatenated operators in terms of data type (where specified)
- Allocation of memory for the pipeline using Fast I/O through the CPDriver
Scheduler
Hospitals use priorities to triage patients appropriately based on severity of symptoms. This concept has been introduced in the Clara Deploy SDK, where studies of higher urgency can be prioritized over processing other studies. Queuing gives the Clara Deploy SDK the resiliency necessary for you to build fault-tolerant hospital-grade systems that meet the needs of future AI.
The NVIDIA Clara platform has a scheduler that is responsible for managing resources allocated to the platform for executing pipeline jobs, and other resources such as render servers. It is responsible for queuing and scheduling pipeline job requests based on available resources. When the system doesn’t have resources to fulfill the requirements of a queued job, the scheduler retains the pending job until enough resources become available.
Model repository
Managing AI models has been a manual process. With the rise of AI, it may only get more tedious. Not only are there different models for different purposes, but there are also multiple model versions that must be maintained over time.
The Clara Deploy SDK now offers management of AI models for instances of NVIDIA Triton Inference Server. The following aspects of model management are available:
- The ability to store and manage models locally through user inputs
- The ability to pull models in from external stores such as NGC
- The ability to create and manage model catalogs
CLI load generator
When developing application pipelines, it is important to be able to simulate expected load. This is the way that you gain the confidence that your hardware and software are architected in ways that can support the estimated load.
The NVIDIA Clara CLI load generator helps simulate hospital workloads by feeding the NVIDIA Clara platform with a serial workload. It enables you to specify the pipeline used to create the jobs, the datasets used as input for the jobs, and other options:
- The number of jobs to create
- The frequency at which to create them
- Type of dataset (sequential or nonsequential)
- Priority
EGX support
NVIDIA Clara is deployable on EGX-managed edge devices for single-node deployments. Using NVIDIA Clara containers and Helm charts hosted in NGC, you can quickly provision a Clara Deploy environment.
Fast I/O integrated with the NVIDIA Clara platform driver
The integrated Fast I/O feature from the Clara Deploy SDK provides an interface to memory resources that are accessible by all operators running in the same pipeline. These memory resources can be used for efficient, zero-copy sharing and passing of data between operators.
Fast I/O allocations can be optionally assigned metadata to describe the resource, such as data type and array size. This metadata and the allocation that they describe can be easily passed between operators using string identifiers.
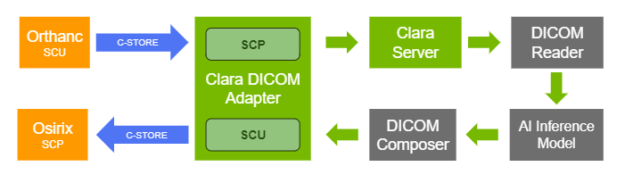
Configuration of the DICOM adapter
Clara Deploy is designed to support different ingestion mechanisms using adapters, depending on workflow needs. It offers a DICOM adapter that can ingest DICOM network objects and facilitate the triggering of jobs based on predefined pipelines with the ingested DICOM objects. The application entities included in the DICOM adapter can be easily configured using RESTful APIs.
Distribution of Clara Deploy in NGC
Getting started with the Clara Deploy SDK has never been easier. The Clara Deploy SDK can now be easily installed over NGC to allow flexible installation options. After the core components are installed, you may pick and choose to install 20+ reference pipelines easily with the NVIDIA Clara CLI.
Reference application pipelines
To help you get started quickly, the Clara Deploy SDK comes with new reference application pipelines to enable your AI workflow approach:
- Detection of COVID-19 in the Chest CT datasets pipeline
- Digital pathology pipeline
- Prostate segmentation pipeline
- Multi-AI pipeline
- 3D image processing pipeline using shared memory
- DeepStream batch pipeline
- DICOM series selection pipeline
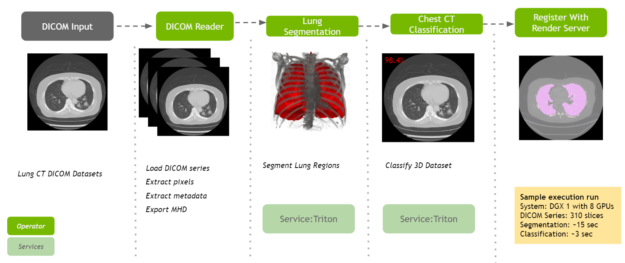
Detection of COVID-19 in the Chest CT Datasets pipeline
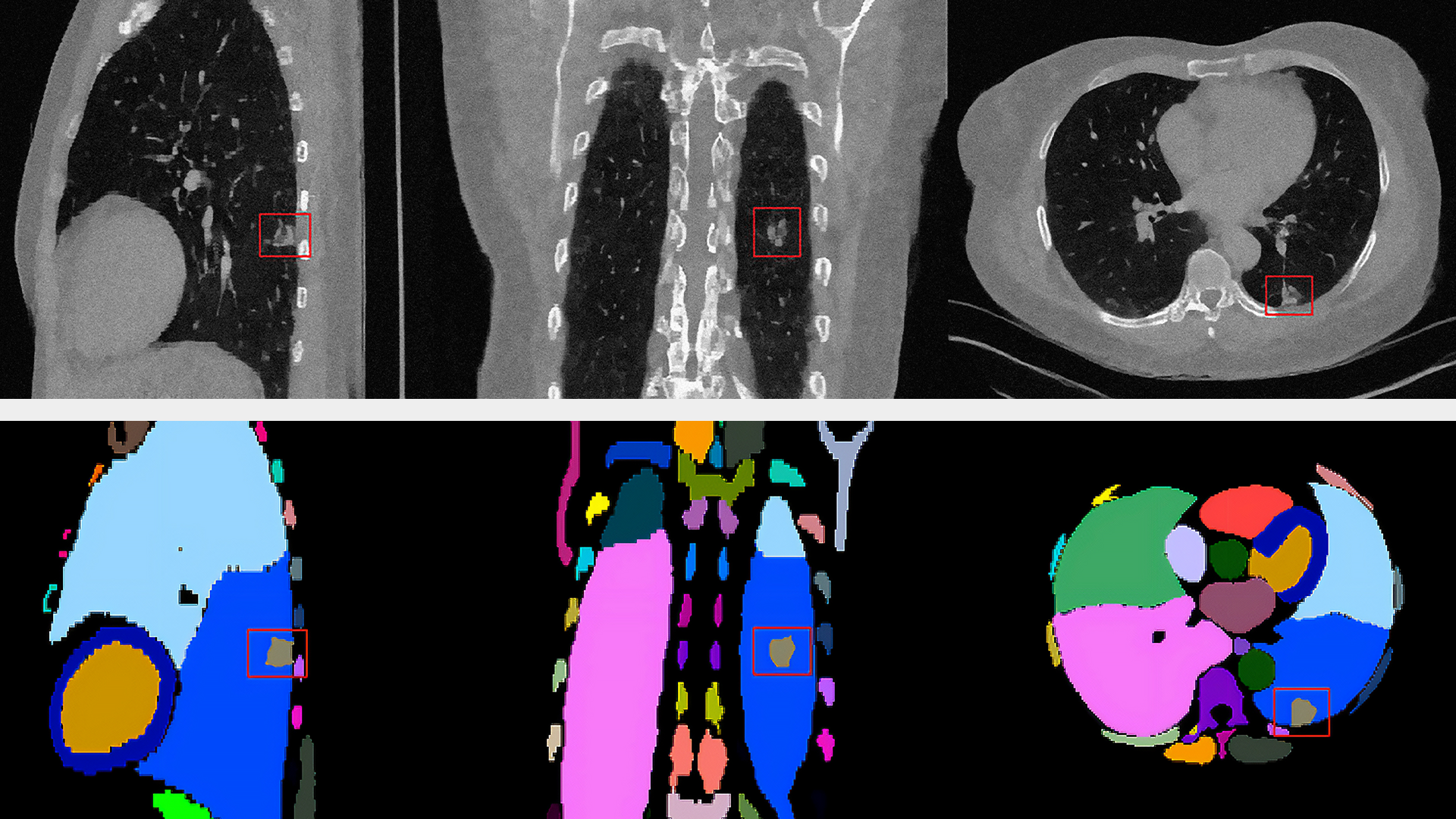
NVIDIA is committed to accelerating and making COVID-19 diagnostic tools available for developers. Clara Deploy offers a reference pipeline that computes the probability of COVID-19 infection in Chest CT DICOM datasets.
It makes use of two new AI operators: the first segments the lung and the second classifies the segmented regions for the presence or absence of COVID-19. The input to the pipeline is a single axial DICOM series of a chest CT scan. The pipeline generates the following outputs:
- A lung segmentation image in MetaImage format
- A new DICOM series for the segmentation image, optionally sent to a DICOM device
- Probabilities indicating COVID-19 and non-COVID-19 in CSV format
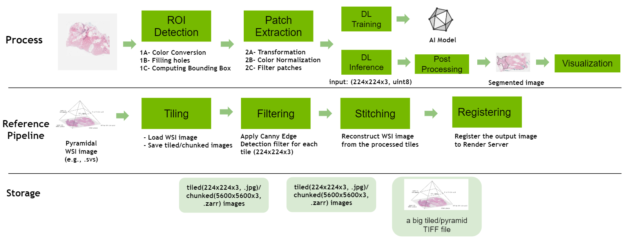
Digital pathology pipeline
The digital pathology features have been expanded to include a new reference pipeline showcasing how large, whole-slide images can be tiled, processed, and stitched together. It contains the following operators:
- Tiling: This operator loads a multi-resolution SVS file, tile it and then writes out the tiled images into disk.
- Filtering: This operator loads each tile from the disk, applies a Canny Edge Detector filter, overlays the resulting edges with blue lines on the original image, and saves the output disk.
- Stitching: This operator loads filtered tiles, stitches them, and saves the resultant image in a multi-resolution TIFF file.
- Registering: This operator sends the final output data to the configured Render Server.
The pipeline features several configurable elements, such as storing the tiled images in JPEG format, or different options to load tiles using the Python Pillow Library, NVIDIA DALI, the Python multiprocessing package and filtering the tiles through the Canny Edge detection filter.
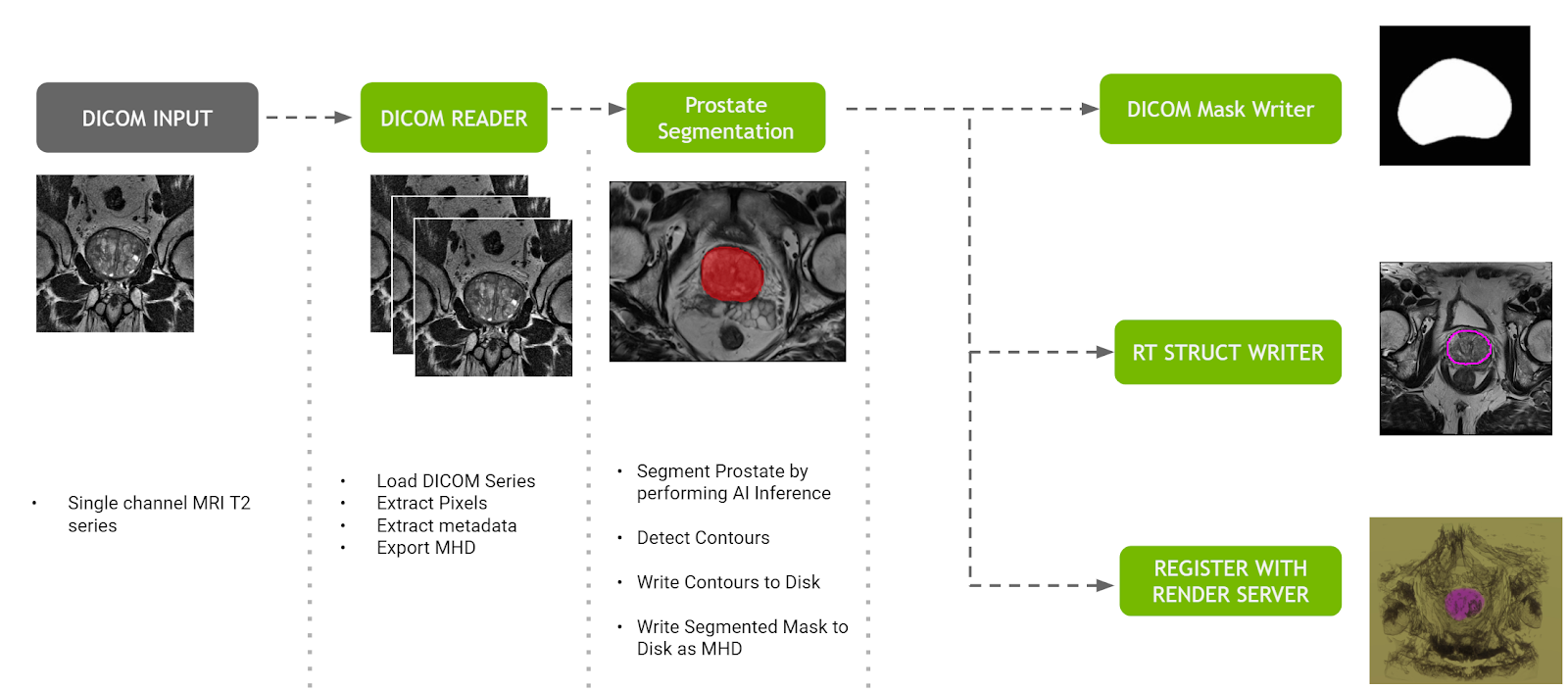
Prostate segmentation pipeline
The prostate segmentation pipeline ingests a single channel MR dataset of the prostate and provides segmentation of prostate anatomy. The pipeline generates three outputs:
- A DICOM RT Structure Set instance in a new series of the original study, optionally sent to a configurable DICOM device.
- A binary mask in a new DICOM series of the original study, optionally sent to the same DICOM device as mentioned earlier.
- The original and segmented volumes in MetaImage format to the Clara Deploy Render Server for visualization on the NVIDIA Clara dashboard.
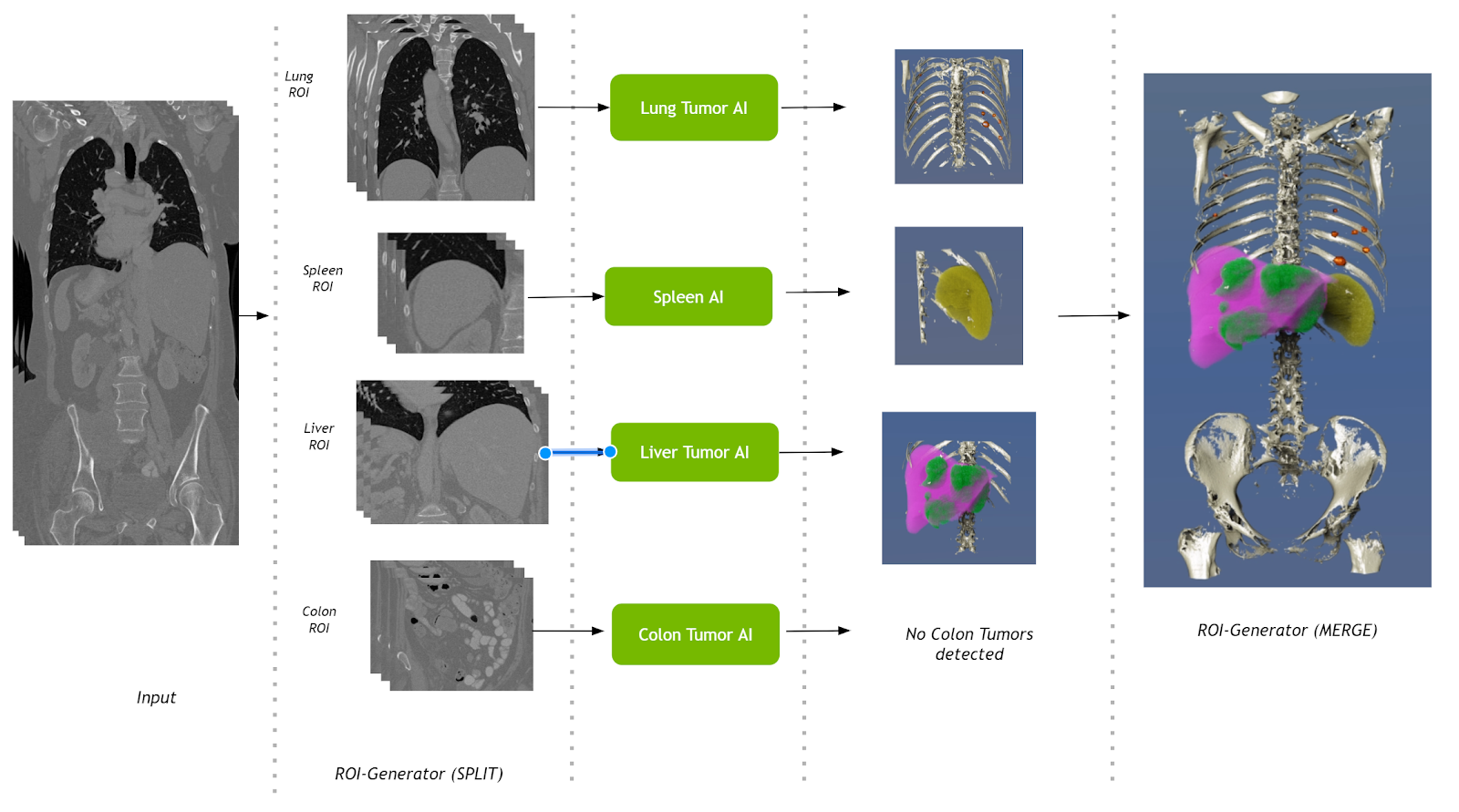
Multi-AI pipeline
This pipeline takes a single CT volumetric dataset as input and splits it into multiple regions of interest (ROIs). These ROIs are then fed into their respective AI operators. Results from the AI operators are finally merged into a single volume. Operators for segmenting liver tumors, lung tumors, colon tumors, and the spleen are used in this pipeline.
The Multi-AI pipeline uses the shared memory–based FastIO interface in the ROI-generator Split operator. The Base AI operator also has been updated with the option to read and write shared memory.
3D image processing pipeline using shared memory
To accelerate the processing of AI pipelines, it is of the utmost importance to keep processes and data in memory, and not cache to disk. Swapping data on and off reduces the performance and ultimately reduces the number of studies that can be performed at any given time. The Clara Deploy SDK provides a reference application pipeline that demonstrates how to leverage shared memory.
The 3D image processing pipeline accepts a volume image in MetaImage format, and optionally accepts parameters for cropping. The output is the cropped volume image and the image is published to the Render Server so that it can be viewed on the web browser. It makes use of shared memory among all operators to pass voxel data around.

DeepStream batch pipeline
The Clara Deploy SDK is used with both medical imaging and videos.
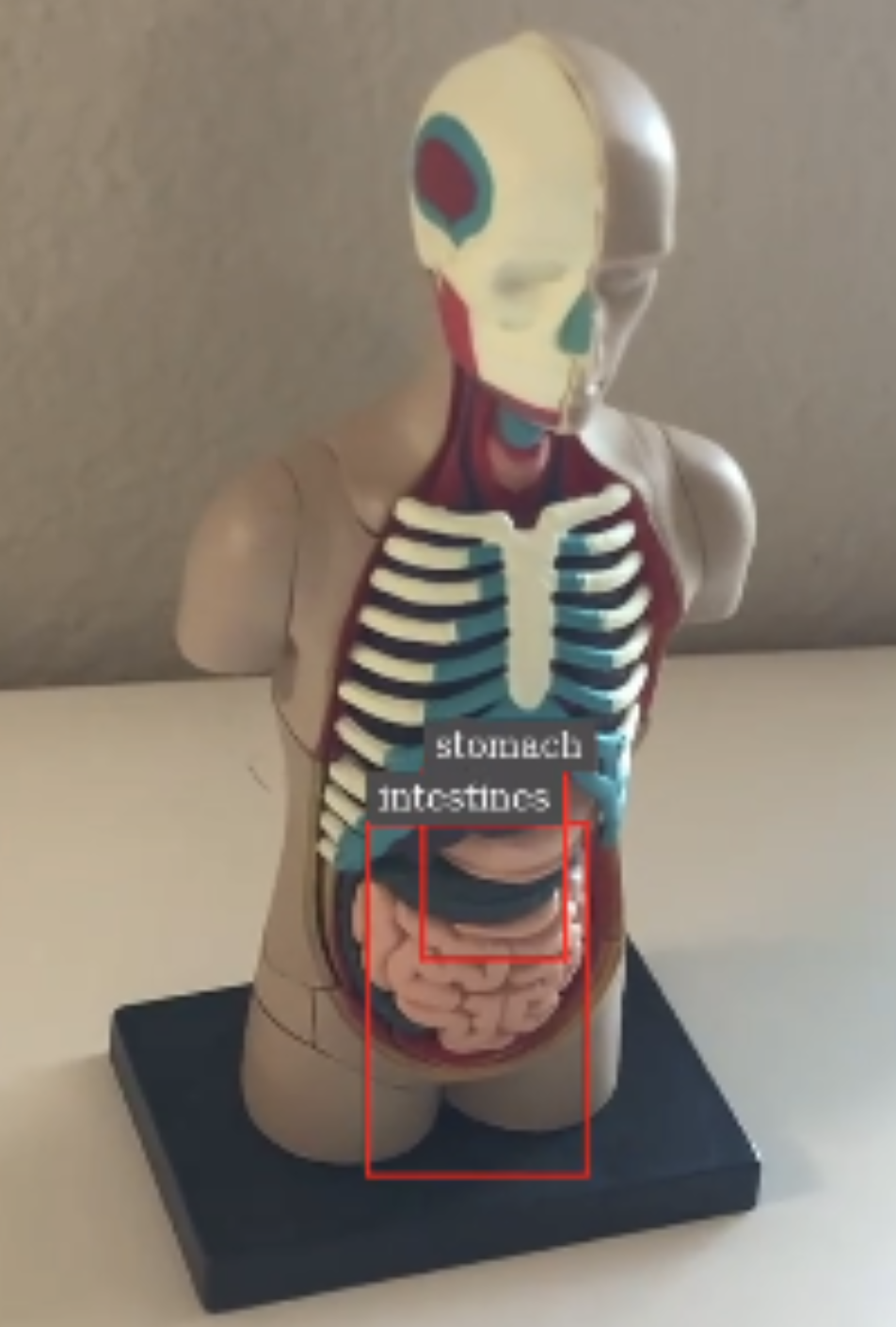
The DeepStream batch pipeline makes use of an organ detection model running on top of the DeepStream SDK, which provides a reference application. It accepts an MP4 file in H.264 format and performs the object detection of finding stomach and intestines from the input video. The output of the pipeline is a rendered video with bounding boxes with labels overlaid on top of the original video in H.264 format (output.mp4), as well as the primary detector output in a modified KITTI metadata format (.txt files).
DICOM series selection pipeline
Medical imaging data passed to Clara Deploy using DICOM can have the headers parsed into JSON format, which can subsequently be used to extract the relevant series with simple matching rules to select the appropriate converted image files.
For example, in a typical CT exam, the first DICOM series acquired is the “scout,” essentially a preliminary image obtained before performing the major portion of a study. However, the scout is only 2D, whereas the CT images in the main series represent a 3D dataset. If an AI model only uses the 3D portion of the imaging study, the scout series can be omitted.
This pipeline can serve as the basis to develop pipelines that can intelligently validate input DICOM studies and choose the correct DICOM series for processing.
New Render Server features
The Render Server, part of the Clara Deploy SDK, provides you with interactive tools to visualize what your AI pipelines are producing. Several key features have been added or expanded in recent releases, including the following:
- Original slice rendering
- Visualization for segmentation masks on original slices
- Oblique multiplanar reformatting
- Touch support for the Render Server
- Multi-resolution digital pathology image support
- Color data type support
- Static scale
- Distance measurement
- Picture-in-picture
Original slice rendering
Not only is it important to see the output of AI processing, but sometimes it is relevant to see the input imaging data. The Render Server can now display the original slices in addition to volume-rendered views.
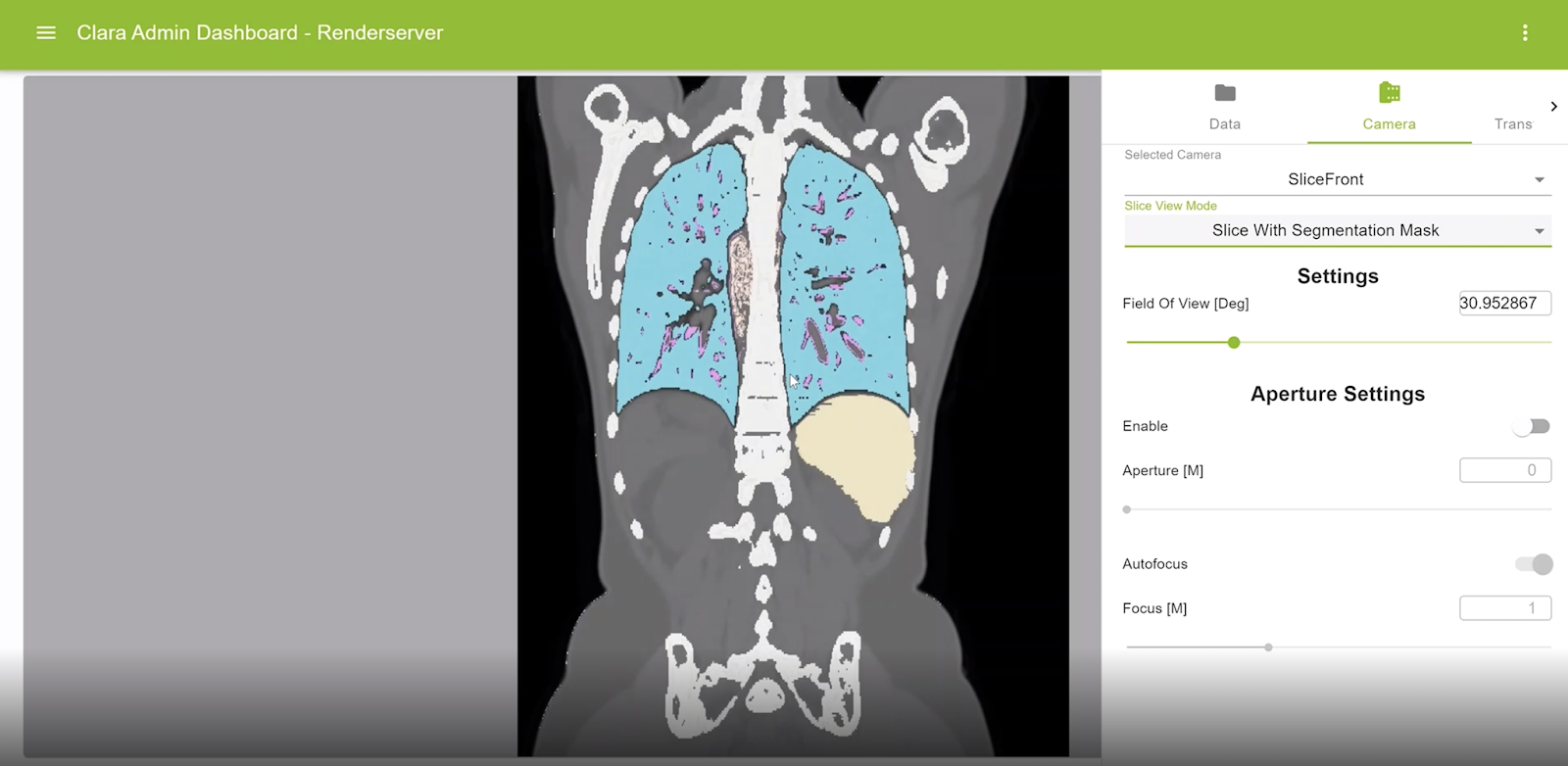
Visualization for segmentation masks on original slices
Segmentation masks can be displayed now on any rendered view of the volume. The color and opacity of such masks are controlled using the corresponding transfer functions.
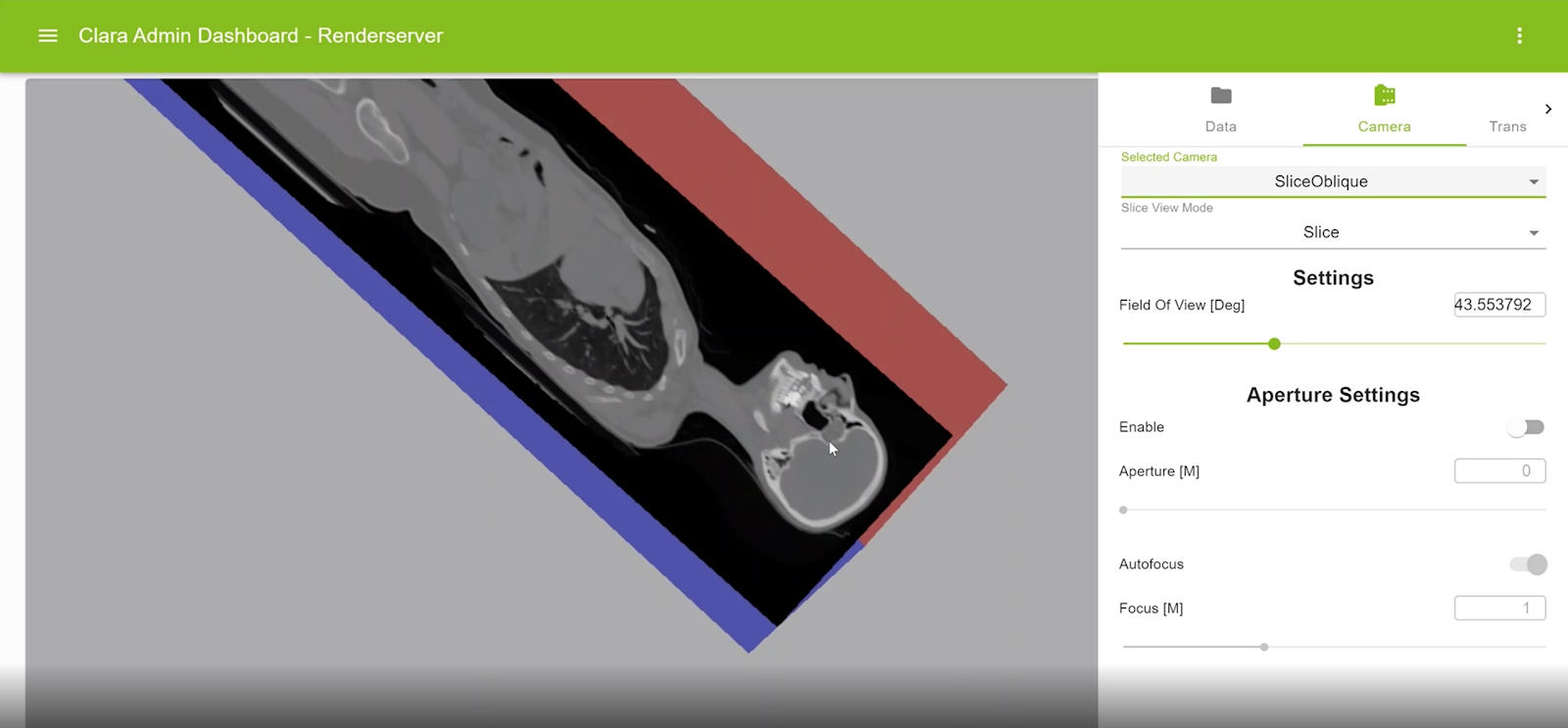
Oblique multiplanar reformatting
This feature enables reformatting the original slices along an arbitrary plane of orientation. For example, axial slices can be reformatted with sagittal or coronal planes. An oblique slice is displayed within the context of a colored axis cube. The view can be rotated, and the displayed slice can be interactively modified.
Touch support for the Render Server
End users may visualize the results of AI processes anywhere. On a touch-friendly device, they can now interact with rendered views using gestures.
Visualization of multi-resolution digital pathology data
Render Server supports loading multi-resolution digital pathology data, which is often encoded in TIFF formats. It can load data at a partial resolution to enables visualization of a large dataset possible even when the system does not have requisite resources to load the highest resolution.
Visible data is loaded on-demand, which makes it possible to view images with a data size larger than the available CPU memory. If a new region of the image needs to be displayed, the image is first scaled up from a coarser level, previously fetched data is then copied for that level, and finally any other missing data is asynchronously fetched.
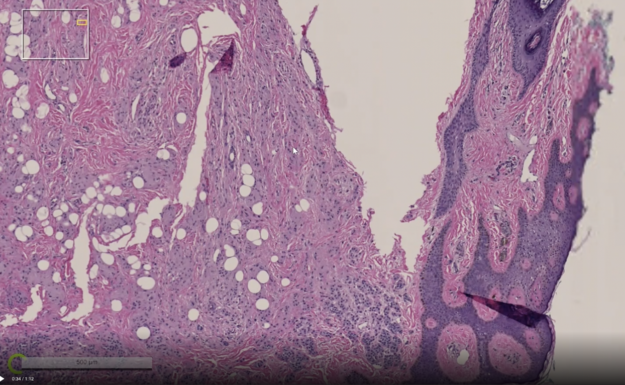
Color data type support
The Render Server interface to configure and specify data has been generalized. You can freely configure data order and dimensions, while density and mask volumes are still supported. Also, there is new support for 2D grayscale and color data with various pixel data types.
Static scale
A scale bar has been added to the bottom left on the Render Server viewer. This enables you to quickly measure the distance between two pixels without specifically creating a line measurement.
Distance measurement
Render Server supports distance measurement on 2D views including original tomographic slices (of modalities such as CT and MR), multiplanar reformatted views, and digital pathology slides. If pixel spacing is included as part of the original data header, the distance between two pixels is provided in physical units.
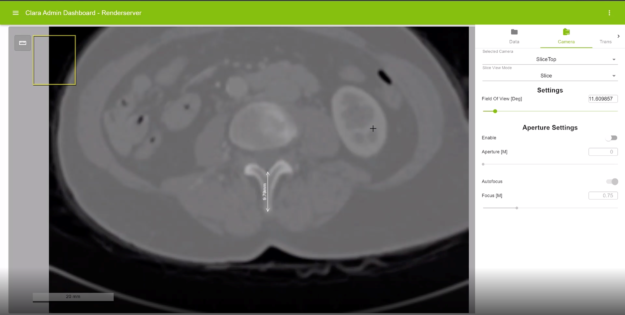
Picture-in-Picture
Render Server is capable of displaying a picture-in-picture view on the top left of the viewer. This view indicates the relative location of the currently displayed region within the whole image.
This feature is especially useful when a large, whole-slide pathology image is displayed on the Render Server. When the user pans or zooms on in a specific subregion of the whole-slide image, this feature can help them identify which portion of the image they are viewing currently.
Management console
A smart hospital that runs hundreds of AI models must have a robust view of all the data being processed at any given time. IT operations, PACS administrators, and even data scientists and model developers benefit from administrative views that allow them to peer into the AI “black box.”
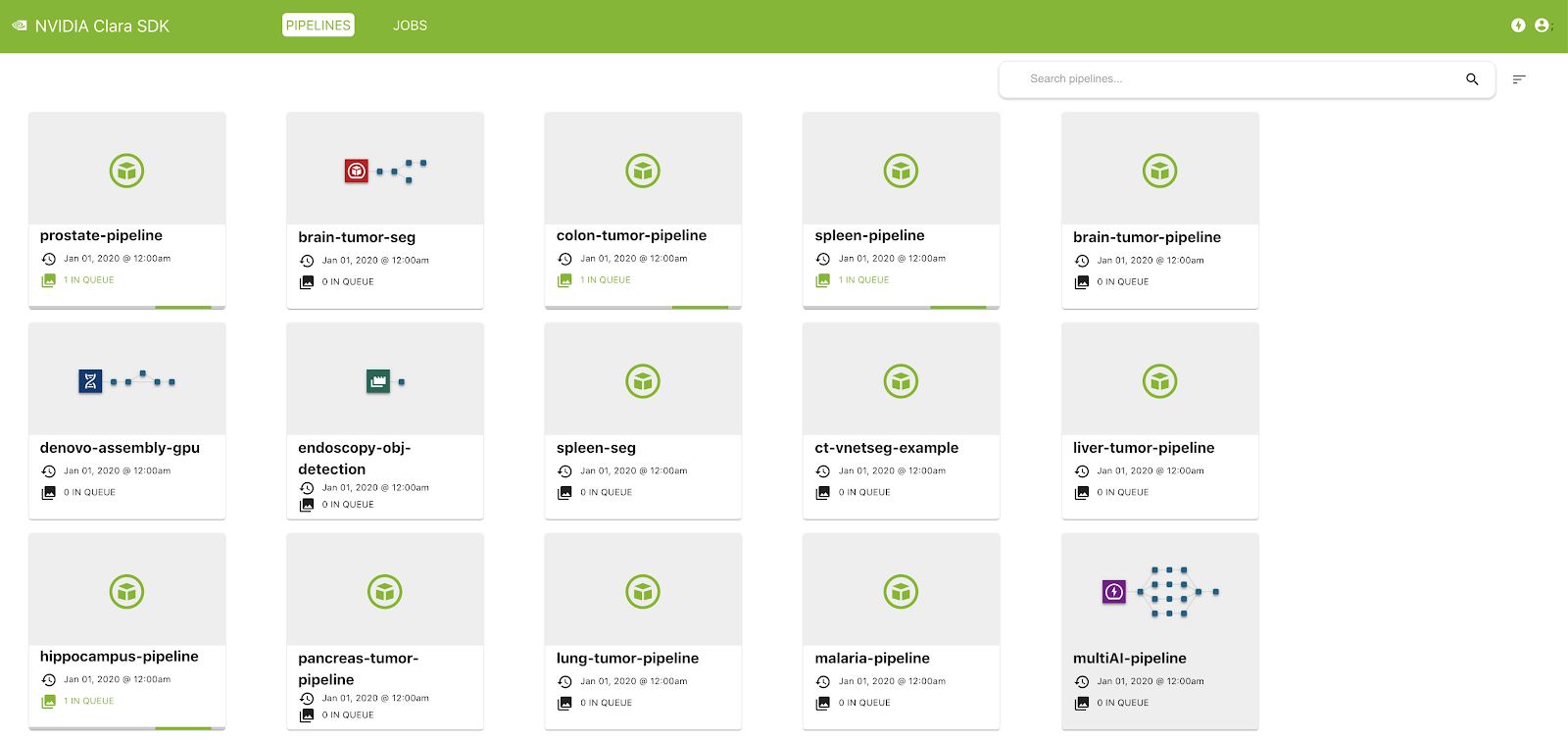
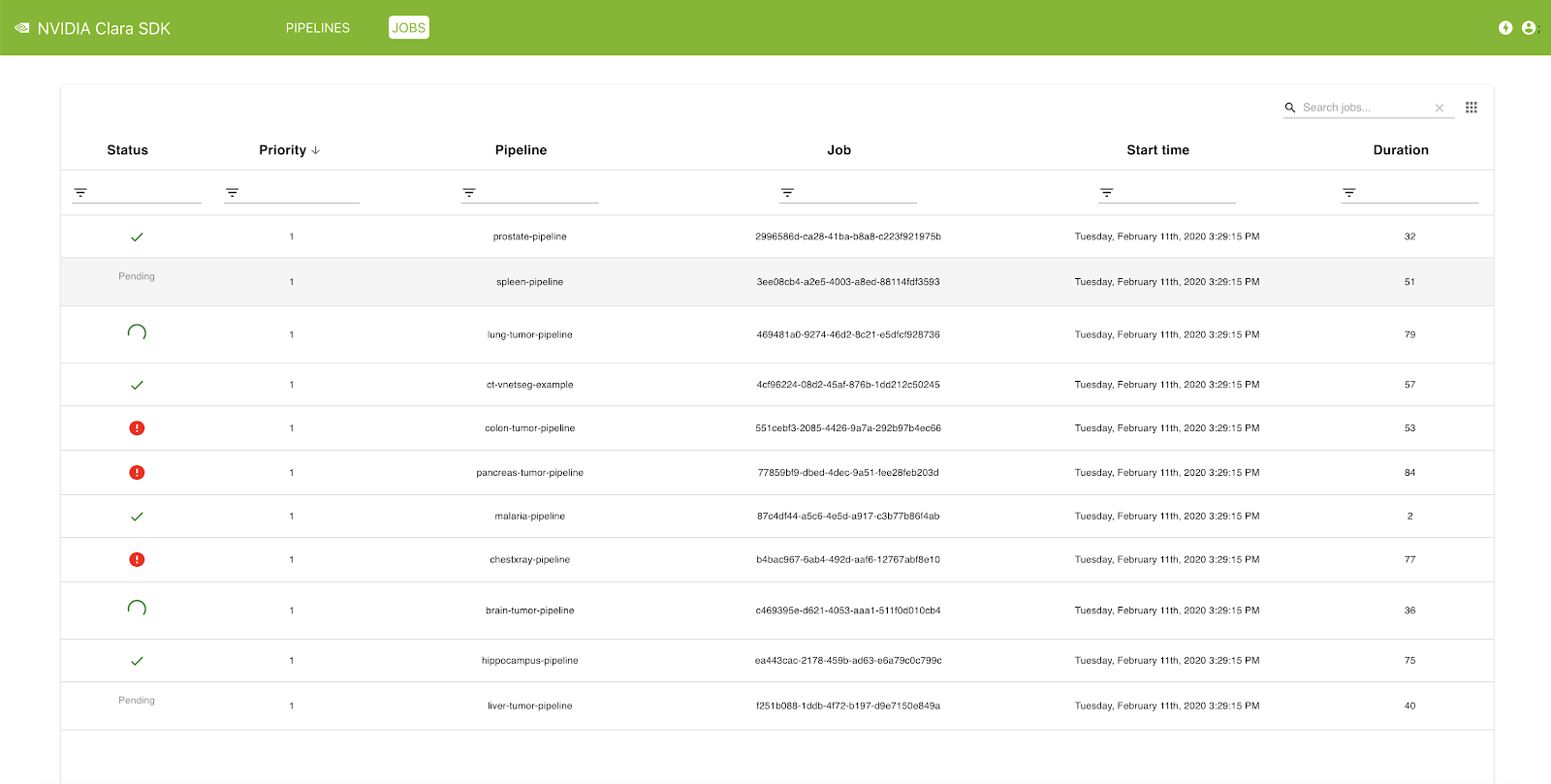
The Clara Deploy SDK offers a management console that can be used to administer pipelines and jobs registered with the Clara Deploy platform. In this release, you can view a list of pipelines with information such as pipeline name, registration date, and the number of jobs queued in the system that were instantiated from this pipeline. Similarly, in the Jobs view, you can see a list of jobs with information such as status, priority, job ID, start time, and duration.
Conclusion
Download the SDK, visit the NVIDIA Clara Deploy SDK User Guide, and view the installation steps. We would like to hear your feedback. To hear about the latest developments, visit the Clara Deploy SDK forum.