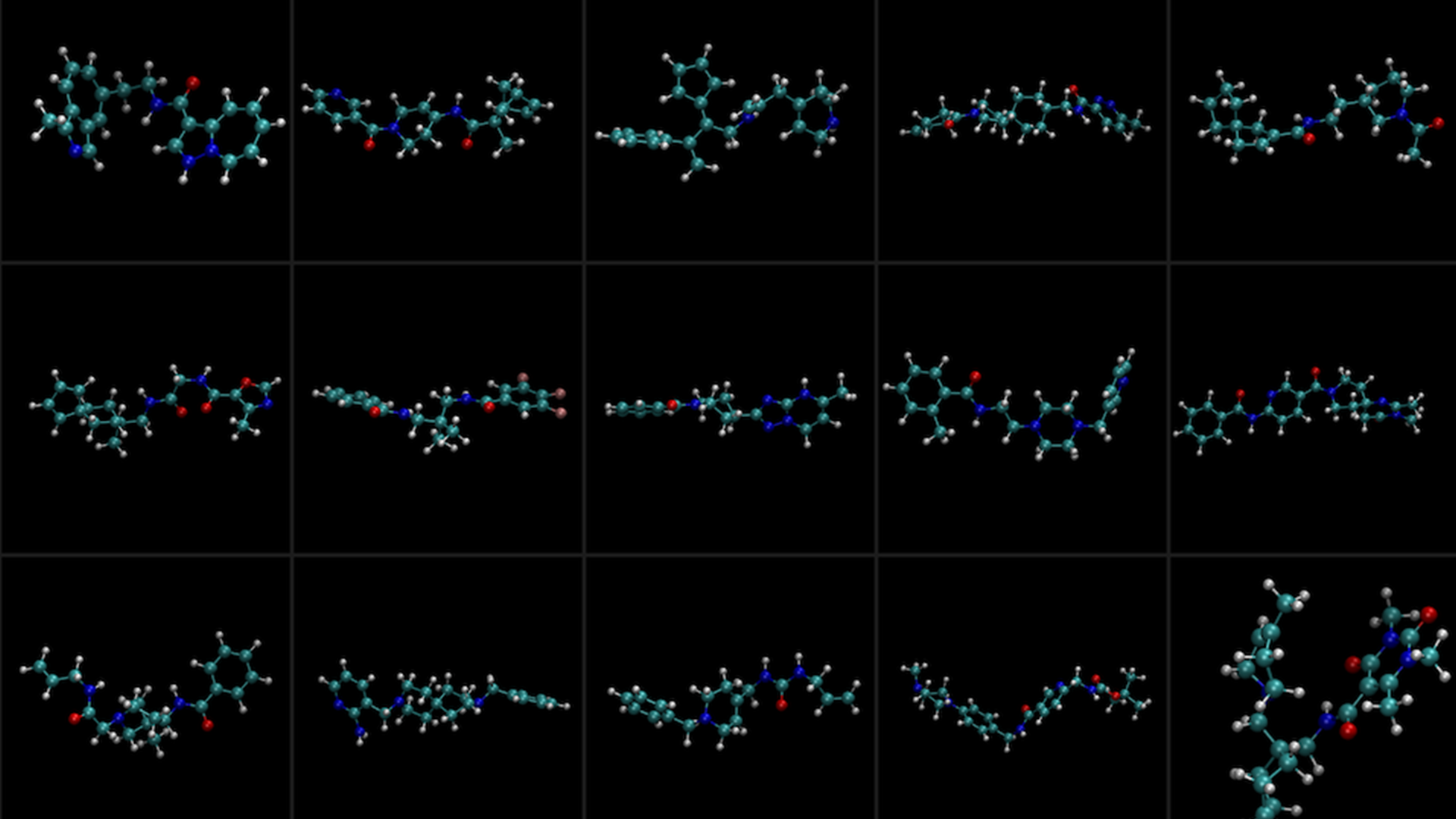
The search for viable drugs is one of the most formidable challenges at the intersection of science, technology, and medicine. Mathematically, the odds of randomly stumbling across a good therapeutic candidate are staggeringly small. This is owed primarily to the astronomically large number of ways that just a handful of atoms can be connected together to make what appear at first glance to be drug-like compounds.
Upon deeper inspection of these molecules, the vast majority would make for unsuitable therapeutics. A clinically viable drug must possess a multitude of characteristics or properties, any one of which—if missing or out of range—could render a drug ineffective or even toxic. Examples of properties that drug hunters seek include those that characterize a drug’s binding affinity, solubility, membrane permeability, molecular weight, and stability, just to name a few.
In essence, the pursuit of drug candidates is a multiple-objective optimization problem.
Generative AI models like NVIDIA BioNeMo’s MolMIM are designed to directly address the challenge of finding molecules with the right properties. Using MolMIM, researchers can generate molecules that maximize a user-specified scoring function, or oracle function for short. MolMIM performs controlled generation, navigating the learned internal representation of chemical space guided by the oracle function that the user provided.
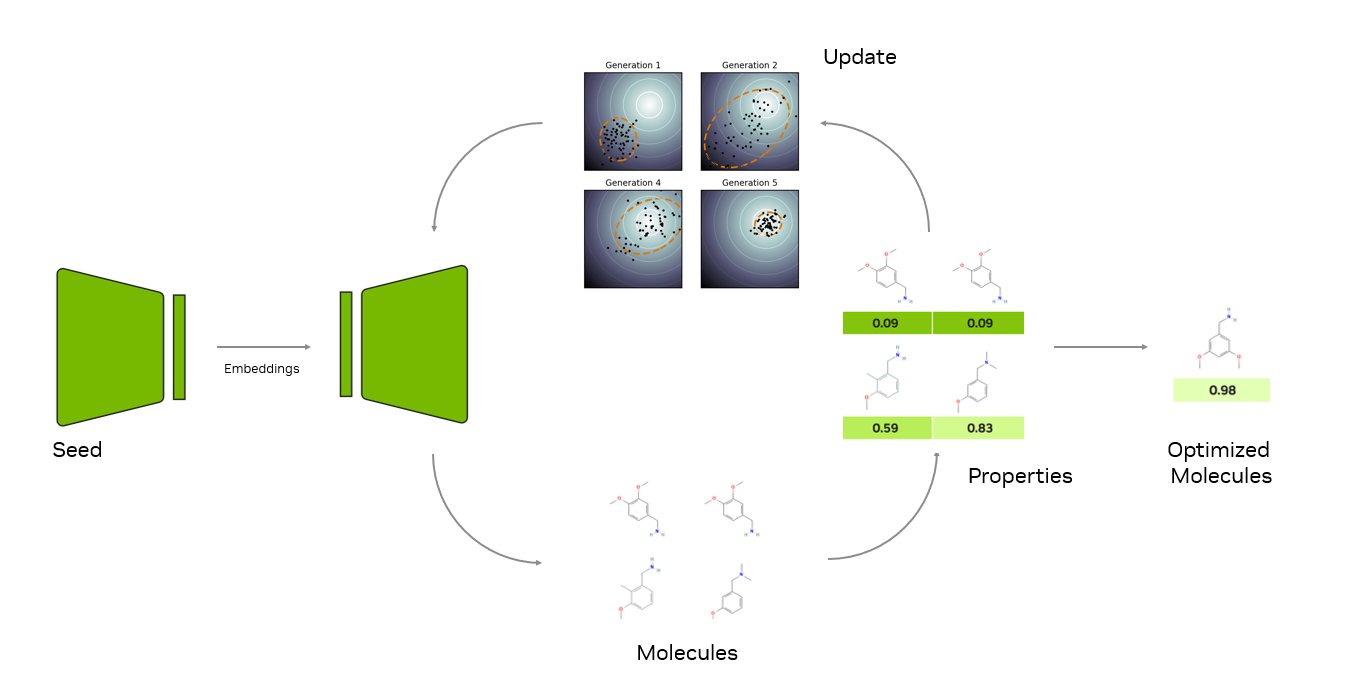
Researchers can define any objective function they wish, even functions that compute multiple objectives. Under the hood, MolMIM uses a gradient-free numerical optimization algorithm called CMA-ES to navigate the latent space of the model. The generative process is iterative:
- At each iteration, MolMIM generates a batch of molecules.
- Armed with the user-provided oracle function, the properties of the molecules are calculated.
- The latent vectors are then updated using the covariance matrix adaptation evolution strategy (CMA-ES) to generate the next batch of molecules.
- The algorithm continues until convergence.
The optimized molecular generation workflow is incredibly simple using the BioNeMo Cloud API with BioNeMo’s Python client library. To define an oracle function, all that you have to do is write a Python function that follows a simple call signature. The function should take a list of SMILES and return a NumPy array of scores of the same length. With the oracle function defined, an optimizer can be instantiated from the BioNeMo Python client and molecule generation can begin!
MolMIM is available in early access through BioNeMo’s Cloud API, which launches January 19, 2024.
Also coming to BioNeMo’s Cloud API is an update accelerating DiffDock, an AI model that predicts the three-dimensional structure of a protein-ligand complex, a crucial step in the drug discovery process. With BioNeMo’s updated version of DiffDock, researchers can predict the three-dimensional pose of a protein-ligand complex more than 2.5x faster than a baseline implementation on identical hardware. For more information, see Build Generative AI Pipelines for Drug Discovery with NVIDIA BioNeMo Service.
Sign up for early access to the NVIDIA BioNeMo Cloud API or get started immediately with BioNeMo Framework for model training.