Deep Learning in medical imaging has shown great potential for disease detection, localization, and classification within radiology. Deep Learning holds the potential to create solutions that can detect conditions that might have been overlooked and can improve the efficiency and effectiveness of the radiology team. However, for this to happen data scientists and radiologists need to collaborate to develop high quality AI algorithms. The NVIDIA Clara AI toolkit, now generally available, lowers the barrier to adoption of AI. The Clara Train SDK, part of the Clara AI toolkit, gives data scientists and developers the tools to accelerate data annotation, development, and adaptation of AI algorithms for medical imaging. We’ll cover the new features of the Clara Train SDK in this release version.
The Clara Train SDK
The Clara Train SDK, running on top of optimized Tensorflow, provides the ability to add AI-Assisted annotation to any medical imaging viewer (annotation tool) and provides pre-trained AI Models and development tools to accelerate creation of AI Algorithms for medical imaging workflows. The Clara Train SDK consists of:
- AI assisted annotation APIs. Advanced features for Auto annotation and interactive annotation.
- Annotation Server. Provides pretrained models to the client application for Transfer Learning
- Unified Python based APIs. Exposes techniques like Transfer learning for training models and provides the ability to train from scratch.
- Pre-trained Models. Based on AH-Net, DenseNet, ResNet, Dextr3D packaged as complete 2D/3D Model Applications for organ based segmentation, classification and annotation.
Key Capabilities
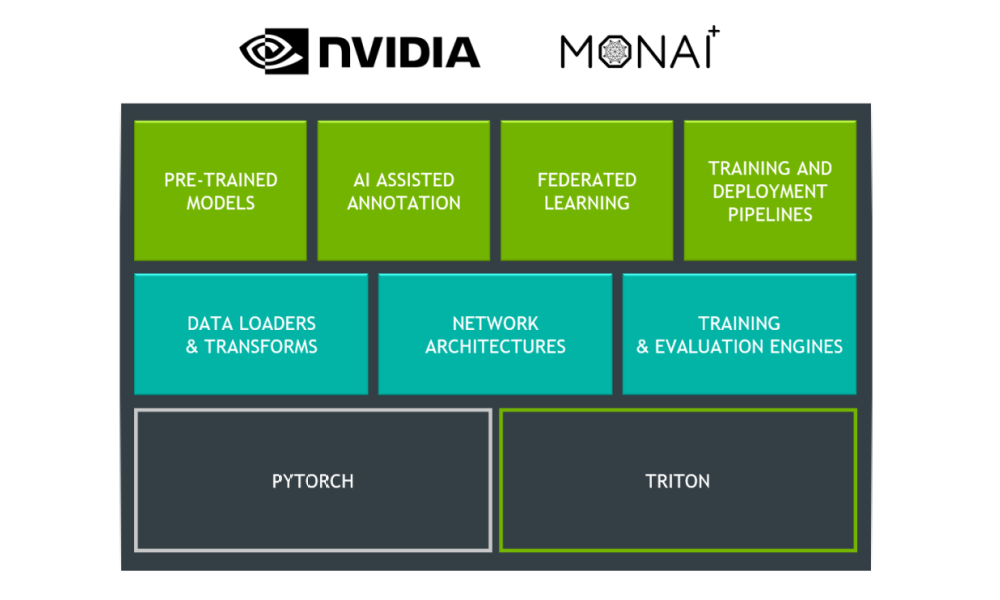
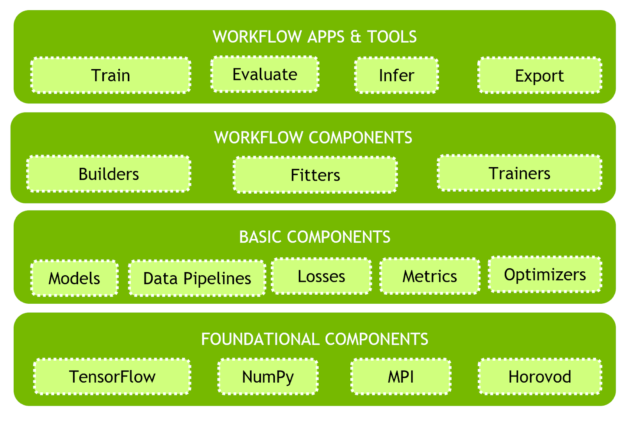
The modular Architecture of Clara Train SDK makes it feasible for data scientists to use functional components or extend additional functionality. It offers an easy-to-use model development environment with useful transforms for data processing pipeline. The development environment includes the building blocks needed to build your AI-assisted medical imaging application, as diagrammed in figure 1.
- Unified Python based APIs. Exposes techniques such as transfer learning for training models and provide the ability to train from scratch.
- State-of-the-art, pre-trained models and training recipes. Packaged as MMARs (Medical Model Archive), providing a unified flexible framework to enable data scientists to bring their own models.
- Ease of integration. For AI assisted annotation APIs.
Modular Architecture
The Clara Train SDK uses a component-based programming model. Deep-learning workflow is comprised of functional components that interact with each other to get the job done.Many implementations for each type of component usually exist. Choosing and configuring components properly allows developers using the SDK more flexible workflow programming.
You’ll find two kinds of components in the SDK:
- Basic components. Includes the principle building blocks of medical deep learning workflows. Examples of such components include data pipelines, model architectures, losses, metrics, inferers, etc.
- Workflow components. When assembled with basic components in a meaningful way enable implementation of workflow functions. Examples of such components include builders, fitters, trainers, evaluators, etc.
Clara Train SDK provides a layer of abstraction with the Workflow Apps and Tools that provides flexibility to make use of any underlying components with ease as well as replacing with their own.
Data Pipelines and Transforms
A data pipeline contains a chain of transforms applied to the input image and label data to produce the data in the format required by the model. Transforms are configured into the data pipeline to produce input data in the format required by the model. The Clara Train SDK provides numerous transforms for various purposes, including loading image data, applying random noise, random flipping, rotating, resizing, and cropping the input image as well as random sampling of positive and negative examples.
Metrics measure the quality of models during the training process. Multiple metric components can be used together to measure different aspects of the model and to find the best model weights dynamically. Tensorboard allows for monitoring the progress of the AI model.
Models: Medical Model Archives (MMAR)
Models provided with Clara Train SDK are packaged as fully developed model applications. Each of these are hosted on NGC – a registry of GPU-optimized AI software including deep learning frameworks, pre-trained AI models and end-to-end industry solutions.
The naming of the application package is based on functionality, organ and modality. For example, segmentation of the liver for CT Images is called segmentation_ct_liver_and_tumor. We took the approach of providing a complete application package in a compressed archive format rather than merely providing checkpoints.
The MMAR (Medical Model Archive) provides a model development environment; defines a standard structure for storing and organizing all artifacts produced during the model development life cycle, as shown in figure 2.
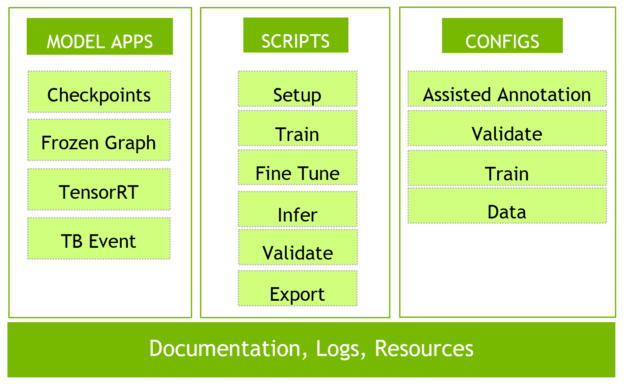
Bring Your Own Model, Transforms, Losses & Metrics
NVIDIA only provided pre-trained models in the early access release. The full release allows users to incorporate their own TensorFlow model architectures and take advantage of the standard framework we have provided. We support any model that follows our Model API spec, such as tf.keras layers. Developers can import the new library provided and extend model class to work to their specific needs. Now, with Clara Train SDK 1.1 we allow researchers to bring their own components, users would write their own components in python file and then point to this file in train_config.json file by providing a path for the new component.
In Transfer Learning Workflow, users download a model package (MMAR), update the config files to point to their datasets, then fine tune them using one of the provided scripts like train_finetune.sh.
In the Training Workflow, users work with the folder structure provided by MMAR and create their own model to be used with the SDK. The organization and structure of an MMAR makes it very easy to construct your own models in Tensorflow with whatever architecture is desired. Figure 3 highlights the application flow.
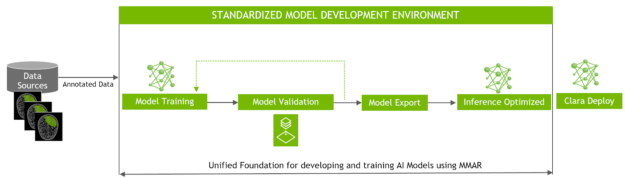
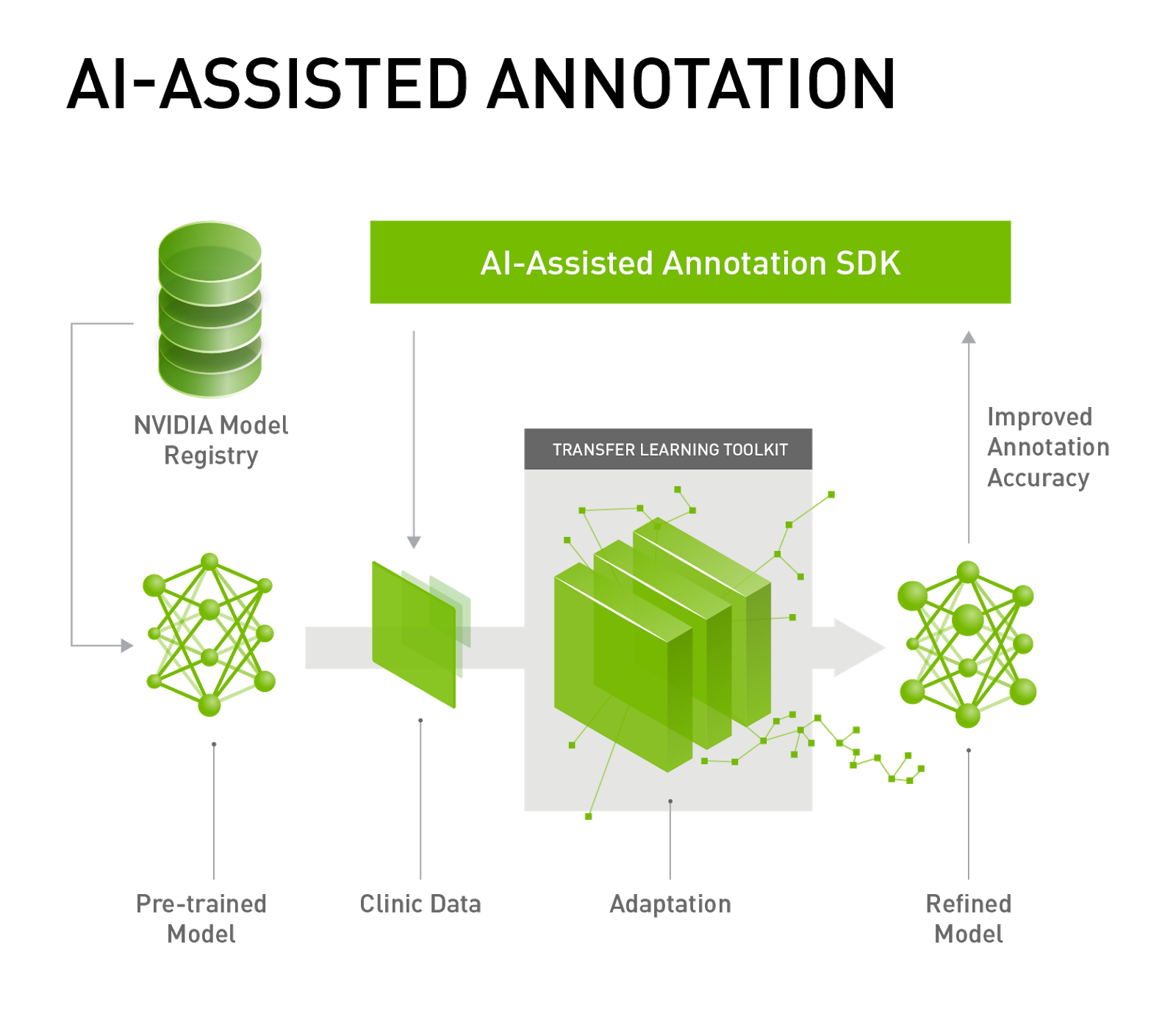
AI Assisted Annotation Enhancements
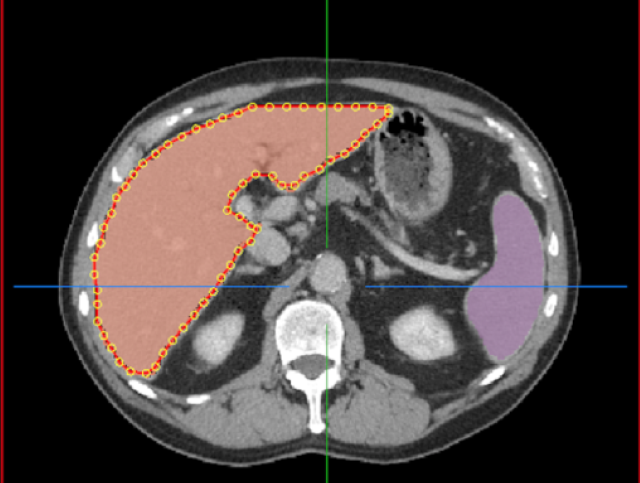
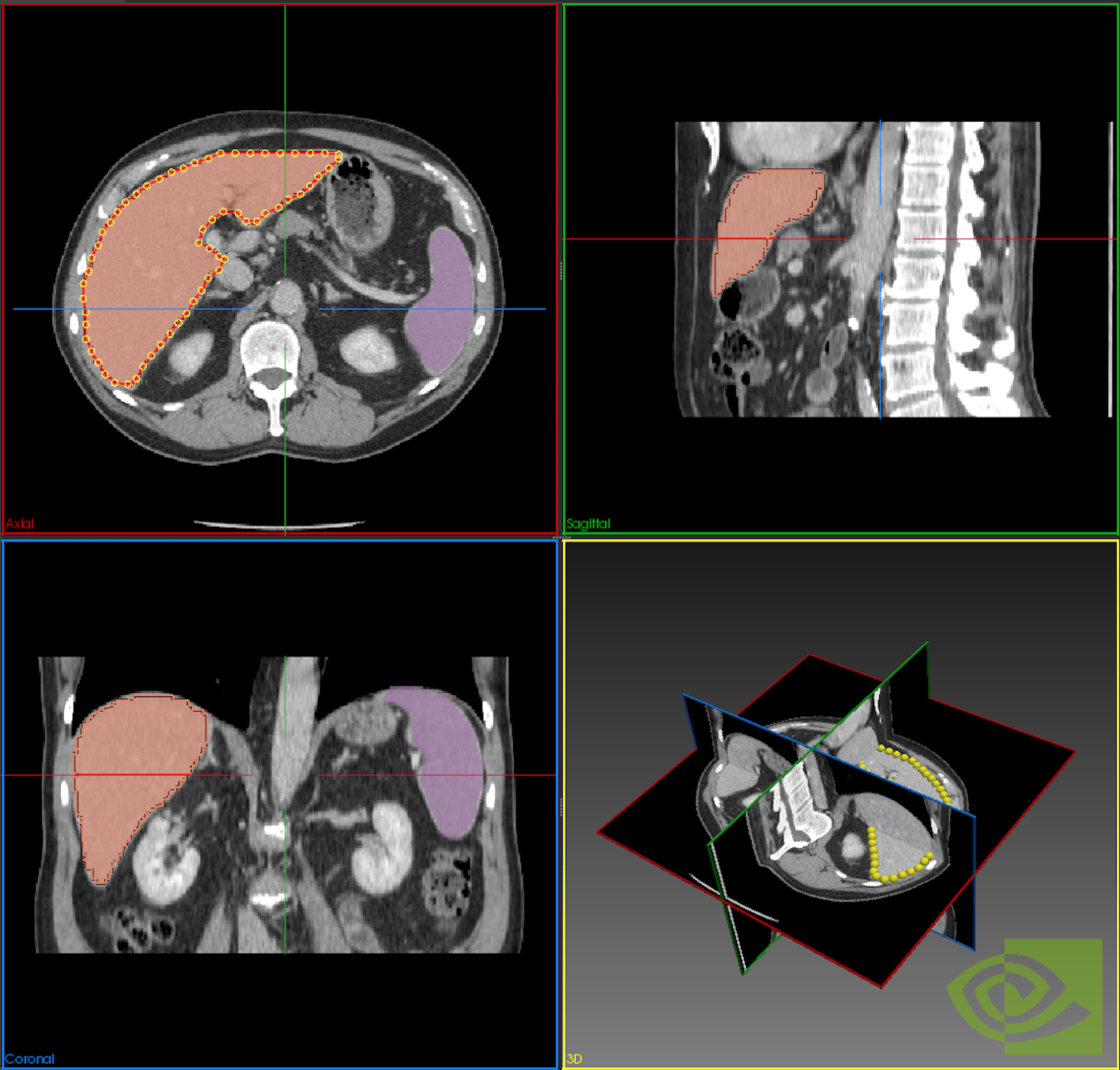
We now provide an Auto Segmentation API which can be used to perform Transfer Learning for an organ segmentation model. Auto Segmentation means users do not need to interactively click points on the image. Instead, users can simply load an image in the viewer and match the label with model loaded in the Annotation Server.
Furthermore, you can now bring your own TensorFlow-trained model to the Annotation Server for Transfer Learning. The Annotation Server and the models needed to automatically annotate various organs can be downloaded from NGC. We also provide the capability for users to easily issue server commands utilizing a web interface, shown in figure 4.
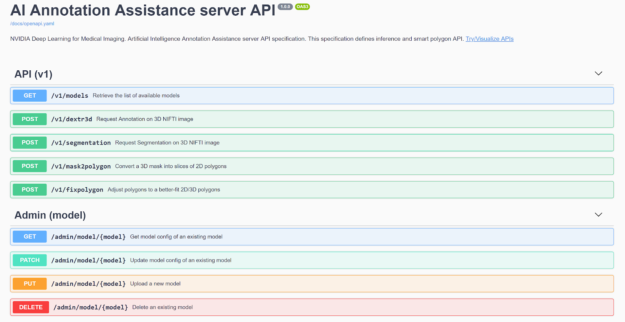
Annotation Workflow
Developers wishing to take advantage of the Clara Train SDK’s full capabilities should work through the following steps:
- Download the Clara Train SDK container from NGC and run it
- Download Models (MMARs) from NGC
- Start the Annotation Server
- Open a Web browser on the port where server is running, check out the documentation
- Select Try and Visualize APIs
- Upload Models to the Server
- Download and Start the reference implementation of AI Assisted Annotation client viewer MITK workbench from Github
- Open Preferences drop down from the MITK viewer and indicate the Server url in ‘NVIDIA Annotation Plugin’ option
- Open image file through MITK viewer; all formats supported by MITK are supported by the plugin
- Choose New label and select the name of organ that is to be labeled
- Select the name of model from Auto Segmentation drop down menu, choose same Annotation Model in the Annotation Section
- Choose Auto Segmentation, at this point Client connects to the Server and runs inference against pretrained model
- The Auto Segmentation result will be used to generate a candidate set of extreme points which the user can adjust and trigger the annotation model
- Make any corrections using the functionality of Interactive annotation and/or Smartpoly polygon editing if necessary.
- Save the newly annotated image
Transfer Learning and Training Workflow
You’ll need to modify the workflow described in the previous section for transfer learning and training as follows:
- In the docker container, open the downloaded model (MMAR) file
- Navigate to the Config folder and choose datalist json file to indicate where your dataset is located and save that file
- Navigate to the commands folder and run the train.sh script. This script trains that model with the dataset specified by user, train_finetune.sh can be used for fine tuning
- Open the included Tensorboard to monitor progress of training
- After training completes, try the validate and infer scripts
- Export model as TensorRT engine to be deployed
Annotate Faster with Clara Train
Get started using Clara Train SDK today. Easily develop, train and adapt your deep learning models.
You can begin with the AI Assisted Annotation Client and Reference Plugin by downloading the client software.The docker Container that includes Python SDK and Annotation Server can be downloaded from NGC. Model Applications can also be downloaded from NGC.
The Medical Imaging Interaction Toolkit (MITK) is a free open-source software tookit by the German Cancer Research Center (DKFZ) for developing interactive medical image processing software. NVIDIA AI-assisted annotation is part of the MITK workbench segmentation plugin and provides 2D and 3D tools for polygon editing and segmentation. Users can download the latest MITK Workbench from the official website. MITK plugin integration source code has been open-sourced by NVIDIA and is avialable in our Github repo.