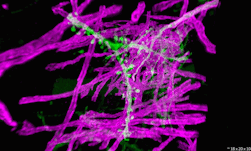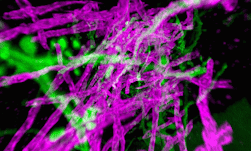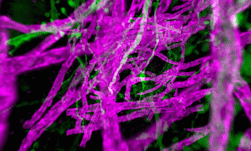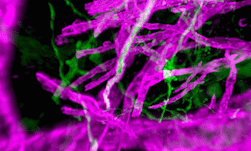
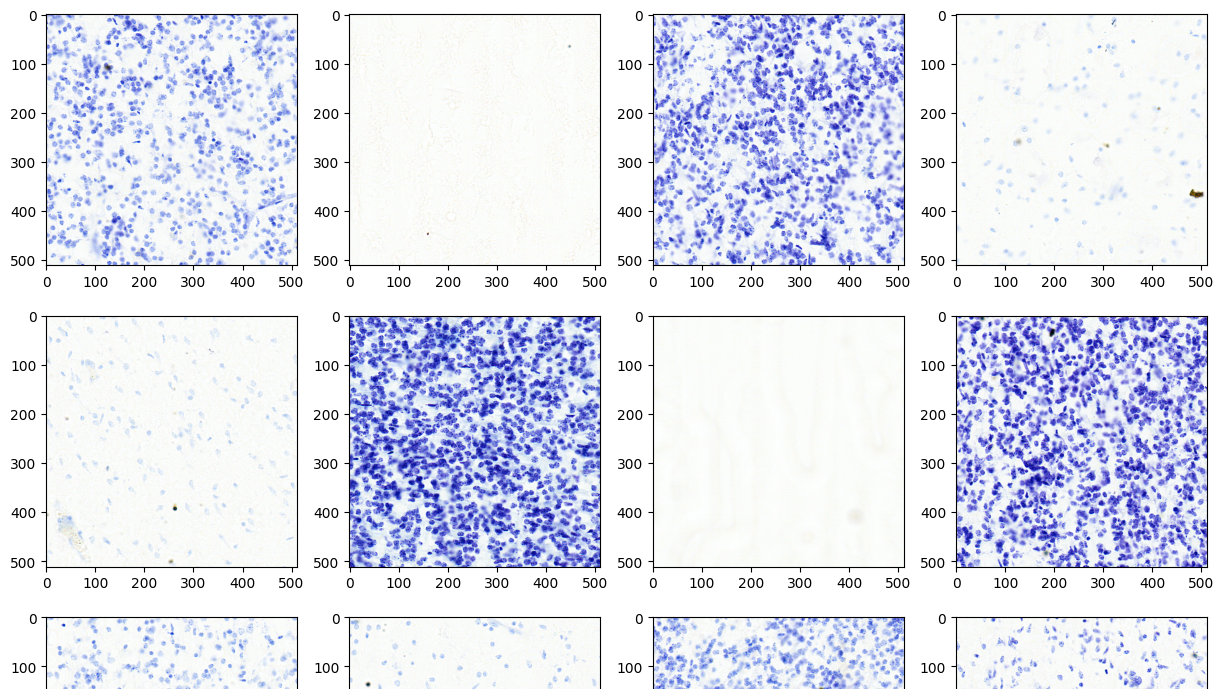
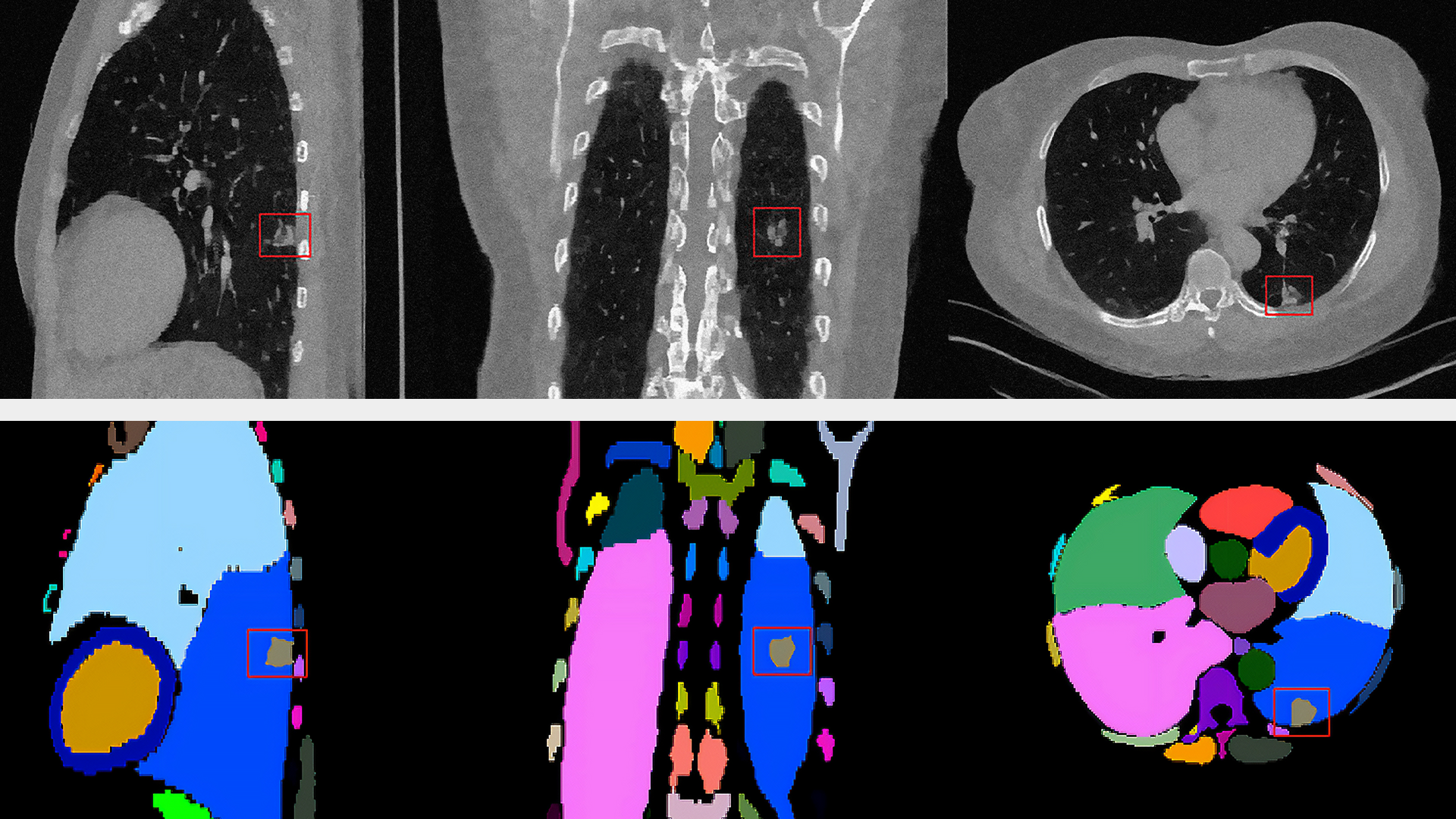
Genomics researchers use different sequencing techniques to better understand biological systems, including single-cell and spatial omics. Unlike single-cell, which looks at data at the cellular level, spatial omics considers where that data is located and takes into account the spatial context for analysis.
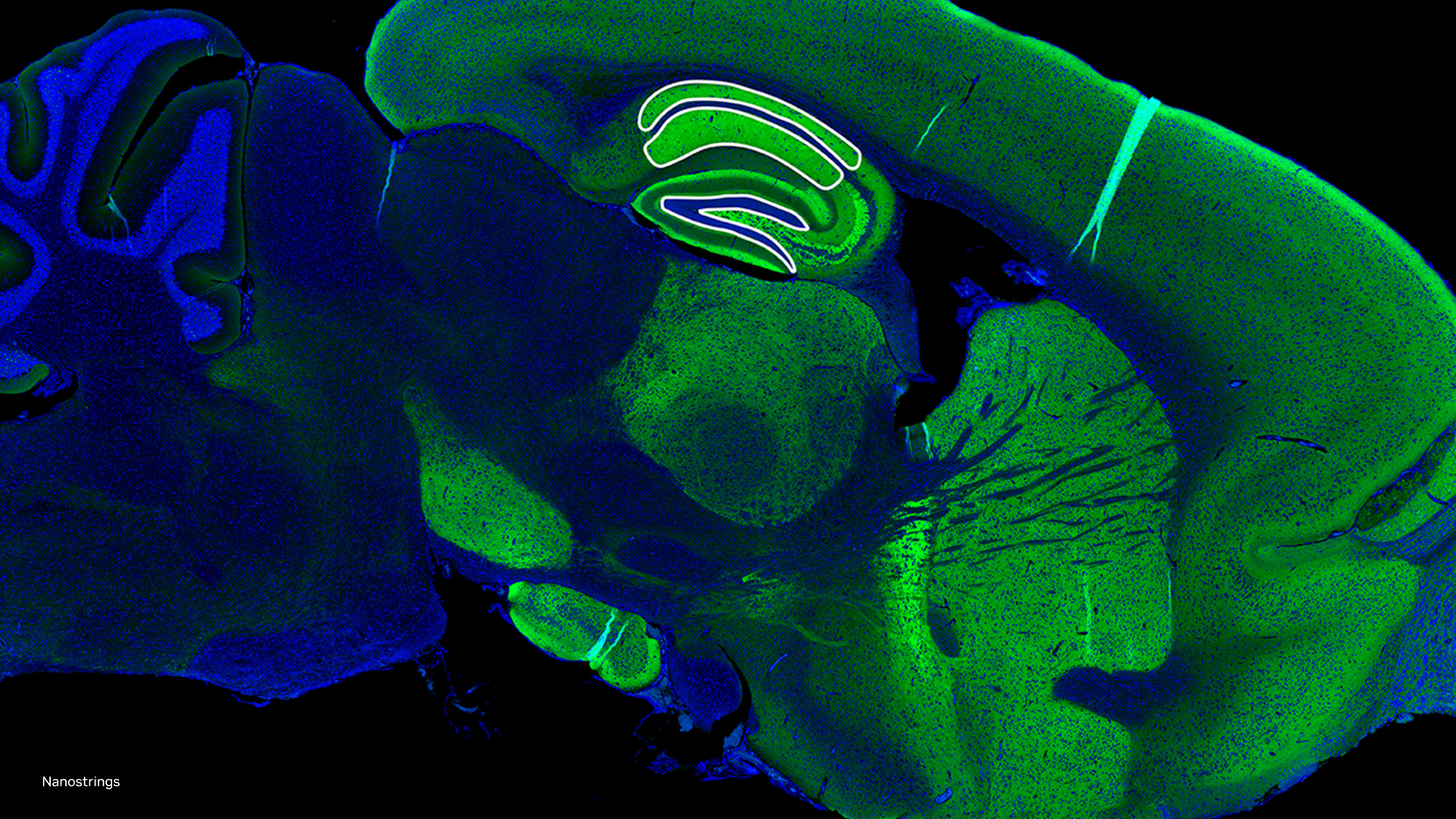
As genomics researchers look to model biological systems across multiple omics at the tissue level, the field of spatial omics is driving a new paradigm in approaches for interrogating cells in spatial context. The majority of these spatial omics approaches rely on imaging, to view markers such as fluorescent tags without dissociation of the cell from the tissue.
These tags can be applied across different molecules (RNA and proteins, for example) and retain the spatial information of the cell from which they originated, and where that cell is located in the tissue. This maturing method to model cellular dynamics is driving new understanding in development and disease, and marks an exciting paradigm shift for researchers.
Spatial omics providers like NanoString are already leveraging NVIDIA GPUs and accelerating the compute onboard their CosMx SMI device to address these challenges. “Spatial-omics technology (CosMx Spatial Molecular Imaging) can now image the entire transcriptome inside cells and tissues, generating data at a density and scale that has never before been accomplished (~150 TB per cm2). This data will play a critical role in transforming our understanding of both health and disease, radically accelerating drug development and spatial diagnostics,” said Joseph Beechem, PhD, chief scientific officer, and senior vice president of Research and Development at NanoString.
“In reality, generative AI will be required to explore the true information content of these images,” he added. “We’re thrilled to continue deepening our partnership with NVIDIA at all levels of the data-to-information pipeline. We invite all of the AI community to join us in this Life Sciences Spatial Biology revolution.”
This post introduces the NVIDIA AI Foundation model for cell segmentation, VISTA-2D, which can be trained on a variety of cell imaging outputs, including brightfield, phase-contrast, fluorescence, confocal, or electron microscopy.
VISTA-2D NVIDIA AI Foundation model for cell segmentation
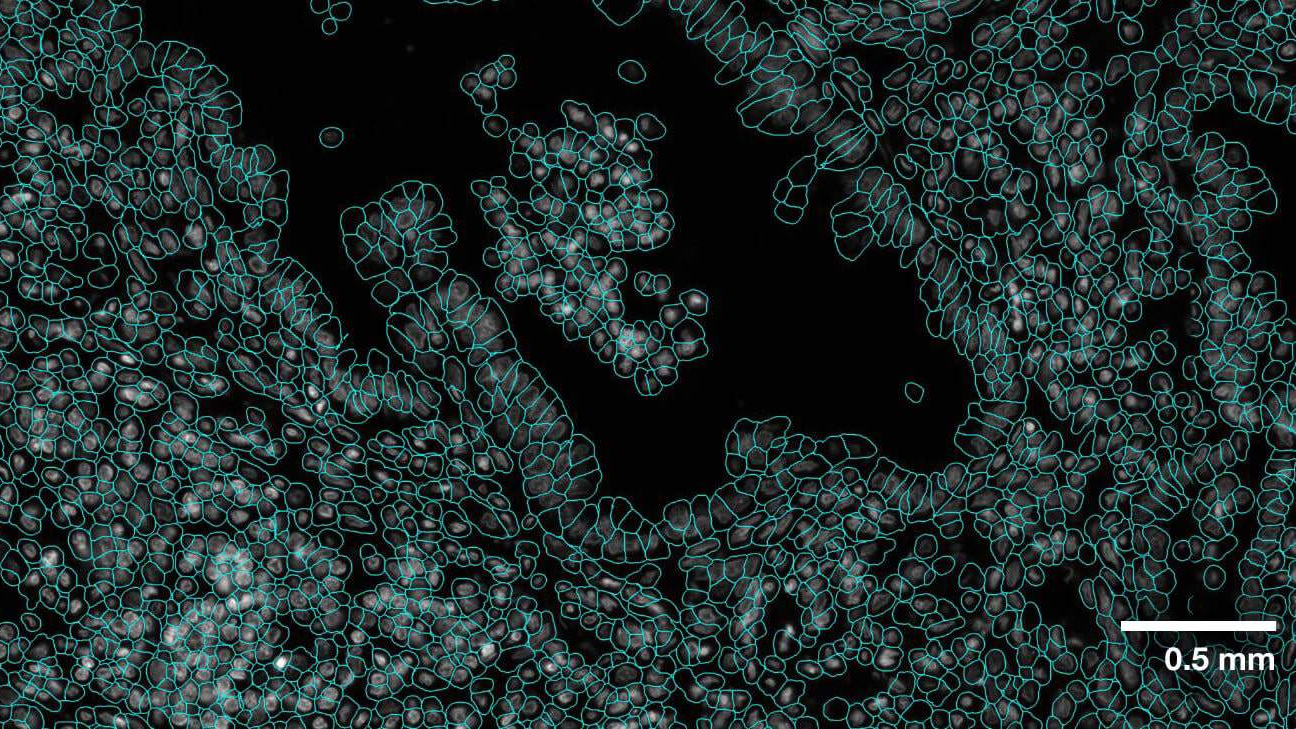
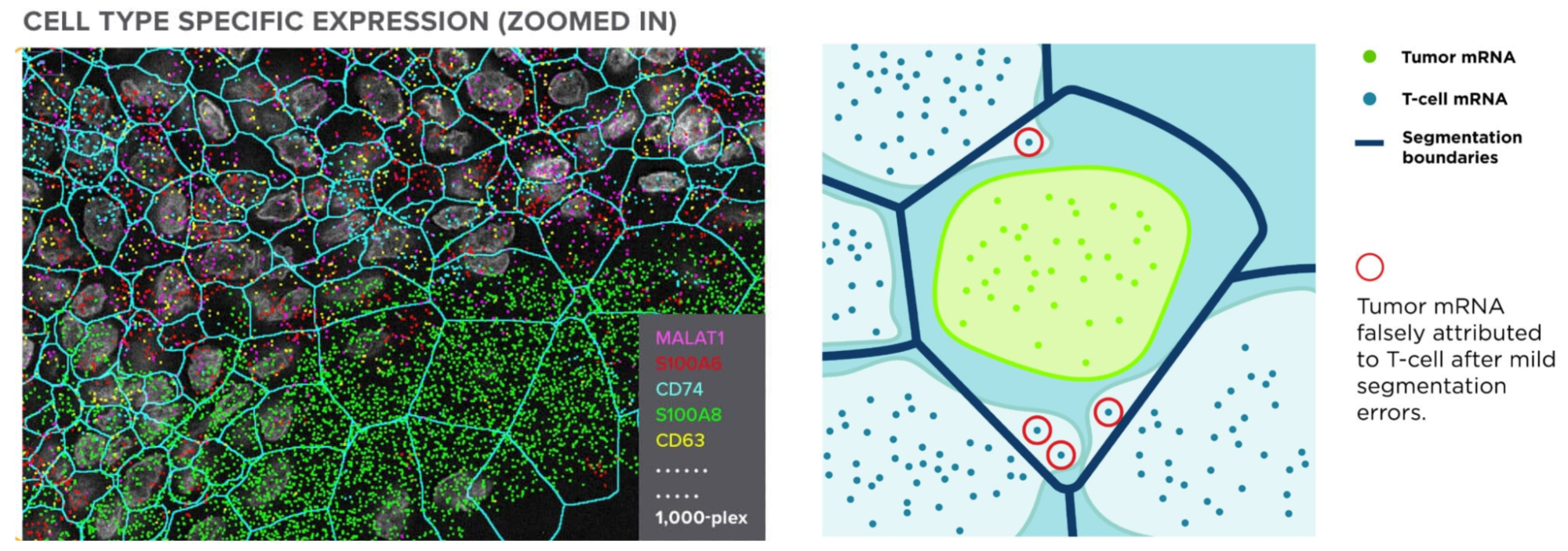
One of the most important steps in analyzing the images of these spatial omics approaches is cell segmentation. As the tags are expressed and counted, attributing those to the correct cell is crucial to receiving accurate results. This relies on the boundaries of the cells having been correctly drawn. Not only is this crucial to all results downstream, but it’s also a nontrivial task requiring recognition of a multitude of different cell types (each with their own morphology), and automation across tens to hundreds of thousands of cells.
VISTA-2D uses a transformer network architecture under the hood, and incorporates Meta’s Segment Anything Model (SAM) pretrained weights to provide a further boost in the performance-based outcome. VISTA-2D is coupled with extensive preprocessing and postprocessing pipelines to enable effective training on any given kind of data. This provides a high-resolution, instance-based segmentation that can be further paired with cell morphology and gene perturbation tasks. VISTA-2D has a network architecture with ~100 million training hyperparameters, making it adaptable and scalable for fast cell segmentation.
Model highlights
- Robust deep learning algorithm based on transformers
- Generalist model as compared to specialist models
- Multiple dataset sources and file formats supported
- Multiple modalities of imaging data collectively supported
- Multi-GPU and multinode training support
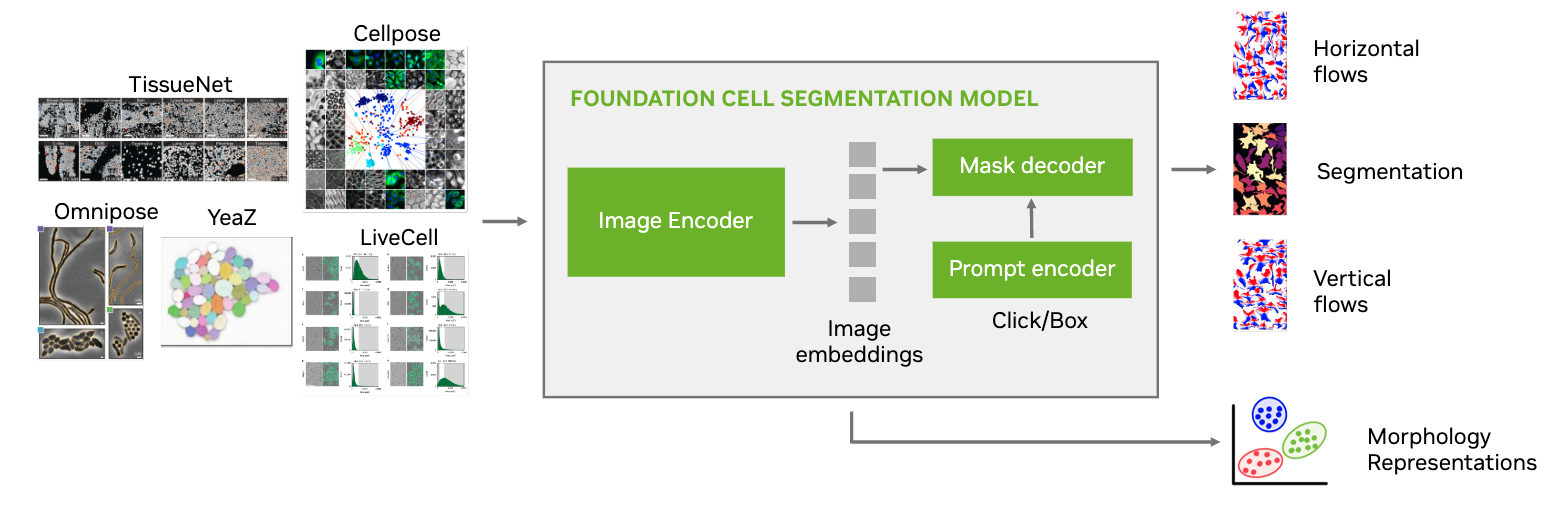
VISTA-2D is a generalist cell segmentation pipeline that is intended to work on multiple types of cells as compared to specialist models that have been proposed in previous literature. The training pipeline is based on the MONAI ecosystem and uses industry-grade code practice. It also includes training support for multi-GPUs that can be scaled to multi-node environments if the training dataset is large enough.
The VISTA-2D training pipeline can also incorporate datasets from multiple sources, different imaging modalities and platforms where the training process is designed to balance the influence of all dataset sources.
The uniqueness of the VISTA-2D pipeline is that it factors in the transformations needed for preprocessing and postprocessing the data, where multiple factors play a role in the performance. For example, vertical and horizontal gradient flows that give the network the ability to disseminate between different instances of a cell and also the shape.
The model is also free of inference-time hyperparameters, which have been found in prior well-known approaches such as CellPose. For example, the need to specify cell diameter size during inference.
Performance on benchmark datasets
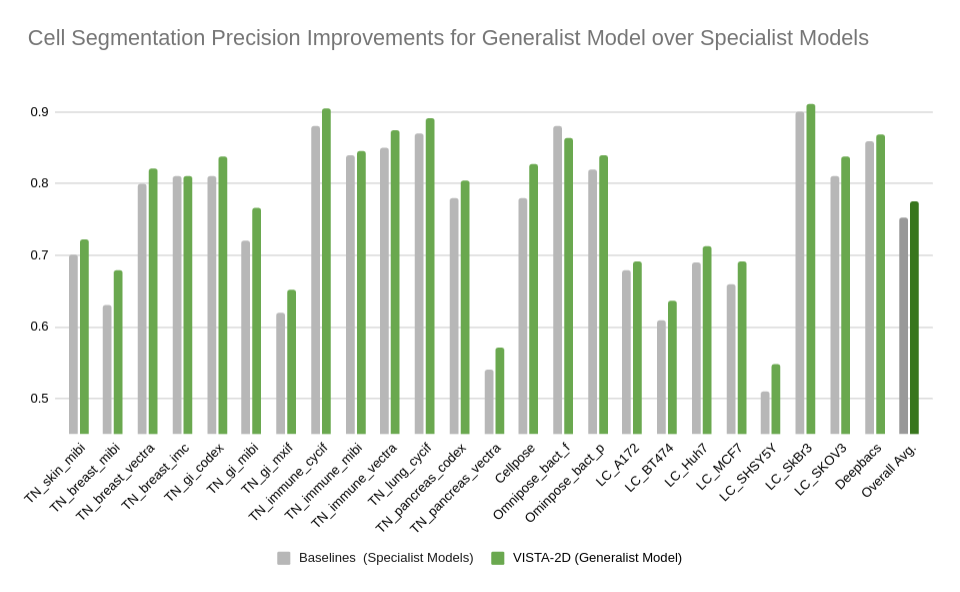
Extensive evaluation was performed for the VISTA-2D model with multiple public datasets, such as TissueNet, LIVECell, Omnipose, DeepBacs, Cellpose, and many more. A total of ~15,000 annotated cell images were collected to train the generalist VISTA-2D model. This ensured broad coverage of many different types of cells, which were acquired by various imaging acquisition types such as brightfield imaging, phase-contrast, fluorescent imaging, H&E, and more. The VISTA-2D model is generalizable and flexible towards all kinds of acquisitions and can be trained on a user’s custom dataset with multichannel support for up to three channels.
The benchmark results of the experiment were performed on held-out test sets for each public dataset that were already defined by the dataset contributors. The well-established standard metric of average precision at an IoU threshold of 0.5 was used for evaluating performance. The benchmark results are reported in comparison with the best numbers found in the literature, in addition to a specialist VISTA-2D model trained only on a particular dataset or a subset of data.
The results indicate that transformer-based network architectures can not only enable state-of-the-art precision when compared to approaches such as Cellpose 2.0, but also train on less data and produce potentially novel insights into morphology of cells through the embeddings produced.
Summary
Cell segmentation is one of the crucial precursor steps to analyzing images obtained from spatial omics techniques. VISTA-2D is an NVIDIA AI Foundation model for cell segmentation that can be trained on brightfield, phase-contrast, fluorescence, confocal, or electron microscopy. With a network architecture of ~100 million training hyperparameters, VISTA-2D is adaptable, fast, and scalable.
To try the VISTA-2D training pipeline using MONAI services, join the early access program.
For more information, check out these NVIDIA GTC 2024 sessions on demand:
- Healthcare Is Adopting Generative AI, Becoming One of the Largest Tech Industries with Kimberly Powell of NVIDIA
- Introduction to GPU-Accelerated Genomics with Parabricks with Harry Clifford of NVIDIA
- First-Ever Whole Transcriptome Imaging of Tissues using CosMx-SMI: Highest-Density Dataset Ever Collected with Joseph Beecham of NanoString