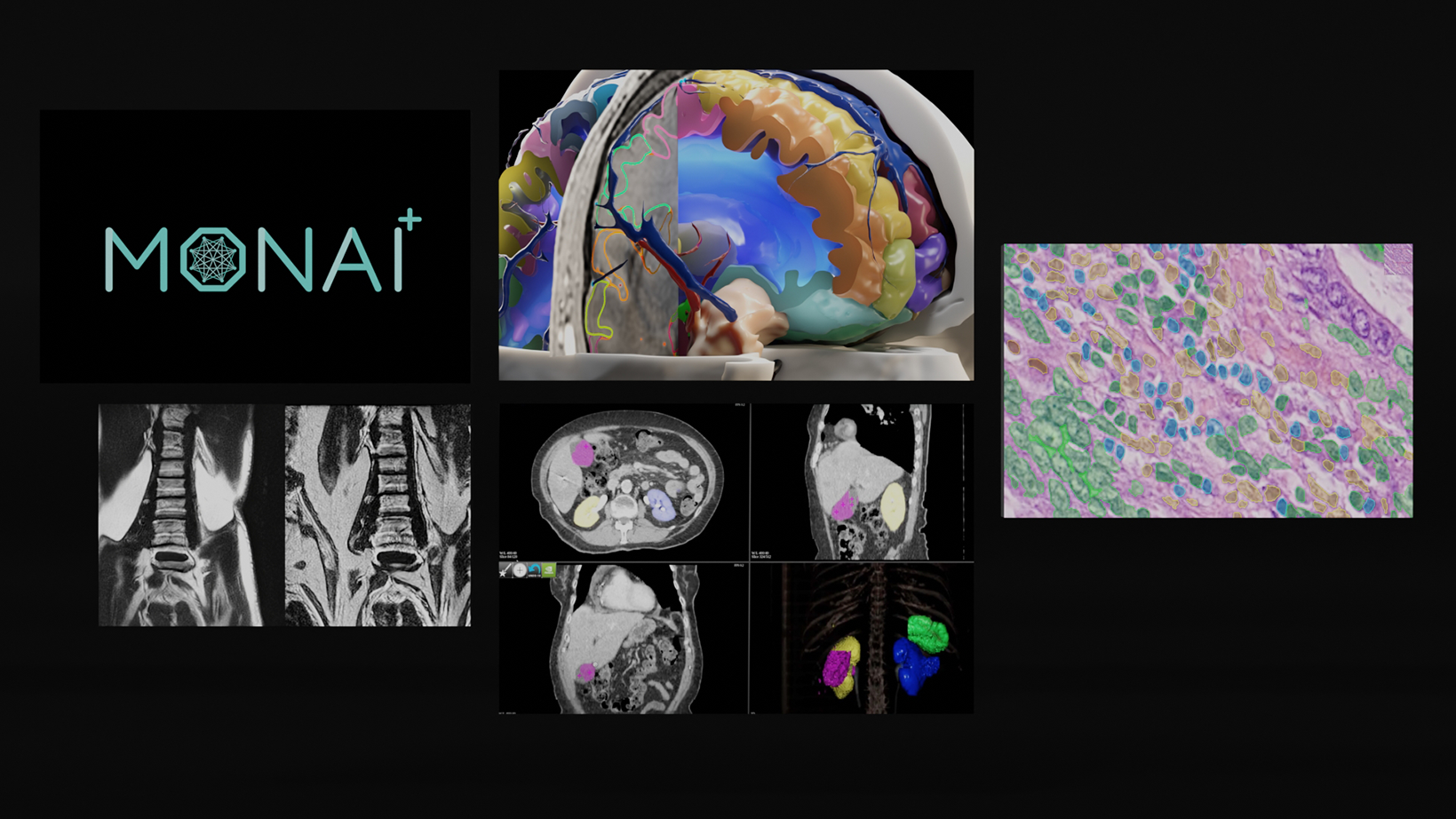
The analysis of 3D medical images is crucial for advancing clinical responses, disease tracking, and overall patient survival. Deep learning models form the backbone of modern 3D medical representation learning, enabling precise spatial context measurements that are essential for clinical decision-making. These 3D representations are highly sensitive to the physiological properties of medical imaging data, such as CT or MRI scans.
Medical image segmentation, a key visual task for medical applications, serves as a quantitative tool for measuring various aspects of medical images. To improve the analysis of these images, the development and application of foundation models are becoming increasingly important in the field of medical image analysis.
What are foundation models?
Foundation models, the latest generation of AI neural networks, are trained on extensive, diverse datasets and can be employed for a wide range of tasks or targets.
As large language models demonstrate their capability to tackle generic tasks, visual foundation models are emerging to address various problems, including classification, detection, and segmentation.
Foundation models can be used as powerful AI neural networks for segmenting different targets in medical images. It opens up a world of possibilities for medical imaging applications, enhancing the effectiveness of segmentation tasks and enabling more accurate measurements.
Challenges in medical image analysis
The application of medical foundation models in medical image analysis poses significant challenges. Unlike general computer vision models, medical image applications typically demand high-level domain knowledge.
Institutes have traditionally created fully annotated datasets for specific targets like spleens or tumors, relying solely on the association between input data features and target labels. Addressing multiple targets is more difficult, as manual annotations are laborious and time-consuming. Training larger or multi-task models is also increasingly challenging.
Despite recent advancements, there is still a long-standing issue in comprehending large medical imaging data due to its heterogeneity:
- Medical volumetric data is often extremely high-resolution, necessitating substantial computational resources.
- Current deep learning models have yet to effectively capture anatomical variability.
- The large-scale nature of medical imaging data makes learning robust and efficient 3D representations difficult, particularly when dealing with heterogeneous data.
However, the modern analysis of high-resolution, high-dimensional, and large-scale medical volumetric data presents an opportunity to accelerate discoveries and obtain innovative insights into human body functions, behavior, and disease.
Foundation models offer the capability to address the heterogeneous variations that complicate the rectification of inter– and intra-subject differences. AI has the potential to revolutionize medical imaging by enabling more accurate and efficient analysis of large-scale, complex data.
A platform for medical visual segmentation foundation models
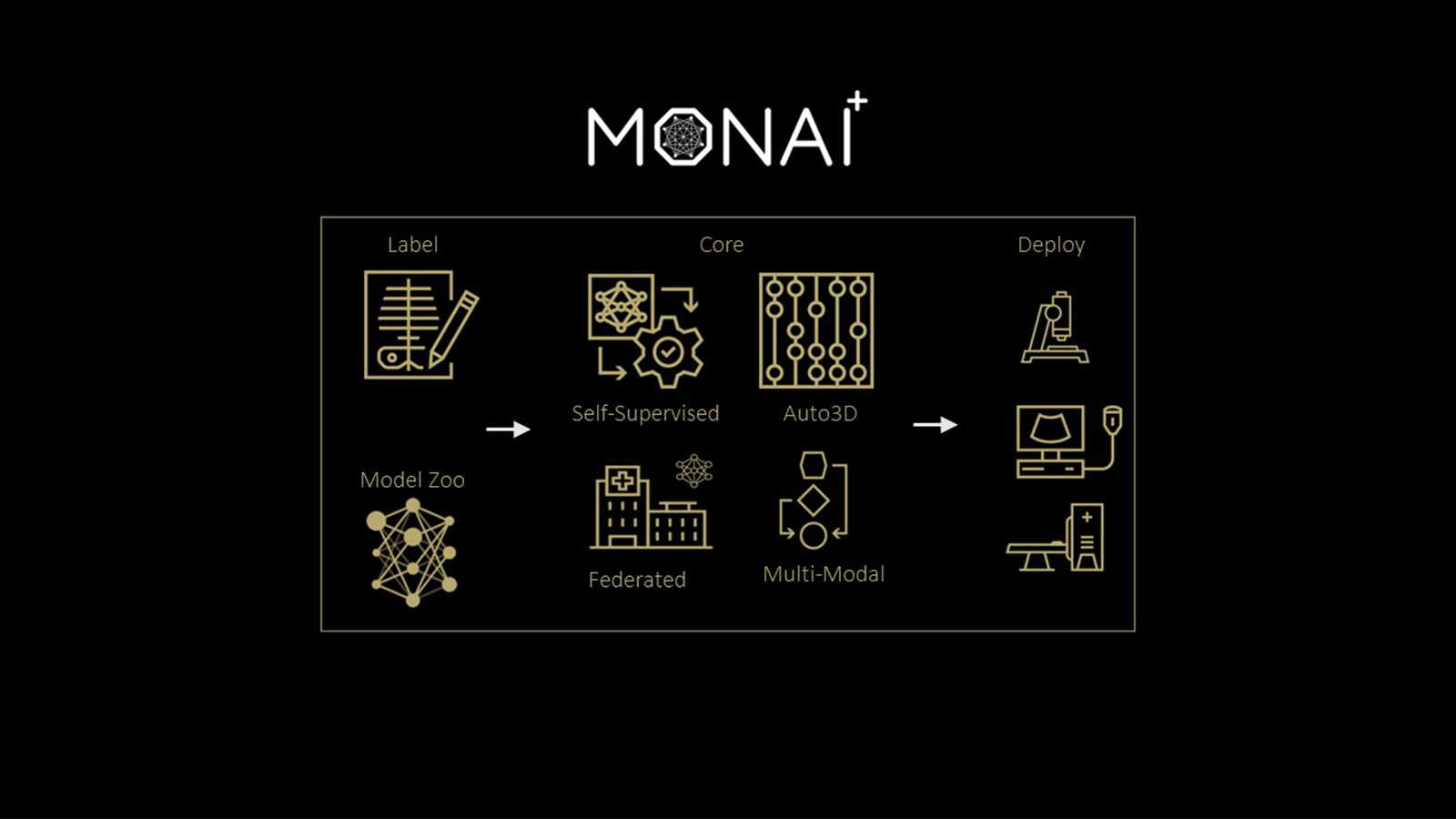
MONAI Model Zoo serves as a platform for hosting medical visual foundation models. It contains a collection of pretrained models for medical imaging tasks developed using the Medical Open Network for AI (MONAI) framework.
The MONAI Model Zoo is a publicly available resource that provides access to a variety of pretrained models for different medical imaging tasks, such as segmentation, classification, registration, and synthesis. These pretrained models can be used as starting points or foundation models for training on new datasets or fine-tuning for specific applications.
The MONAI Model Zoo is designed to accelerate the development of new medical imaging applications and enable researchers and clinicians to leverage pre-existing models and build on top of them.
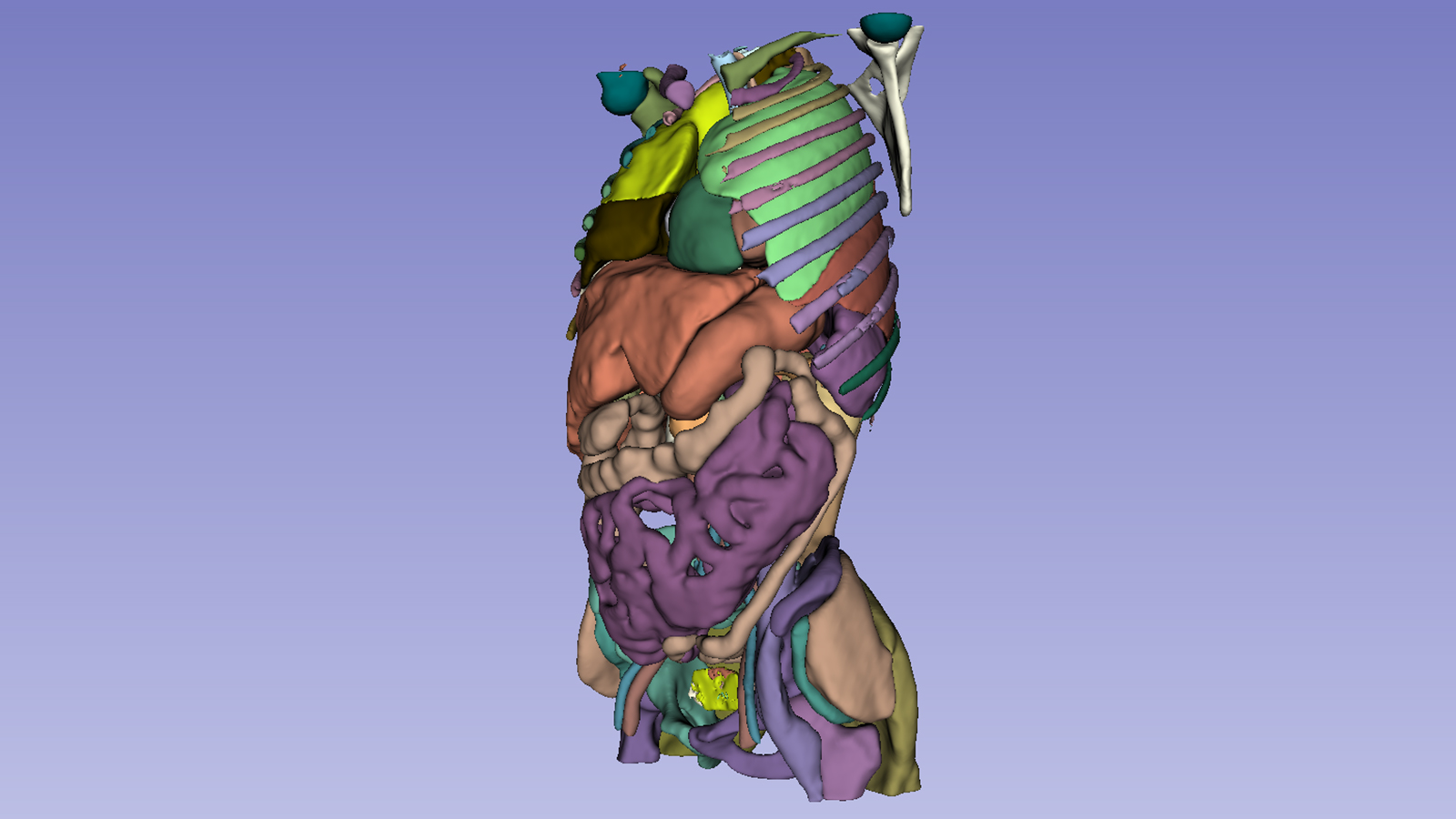
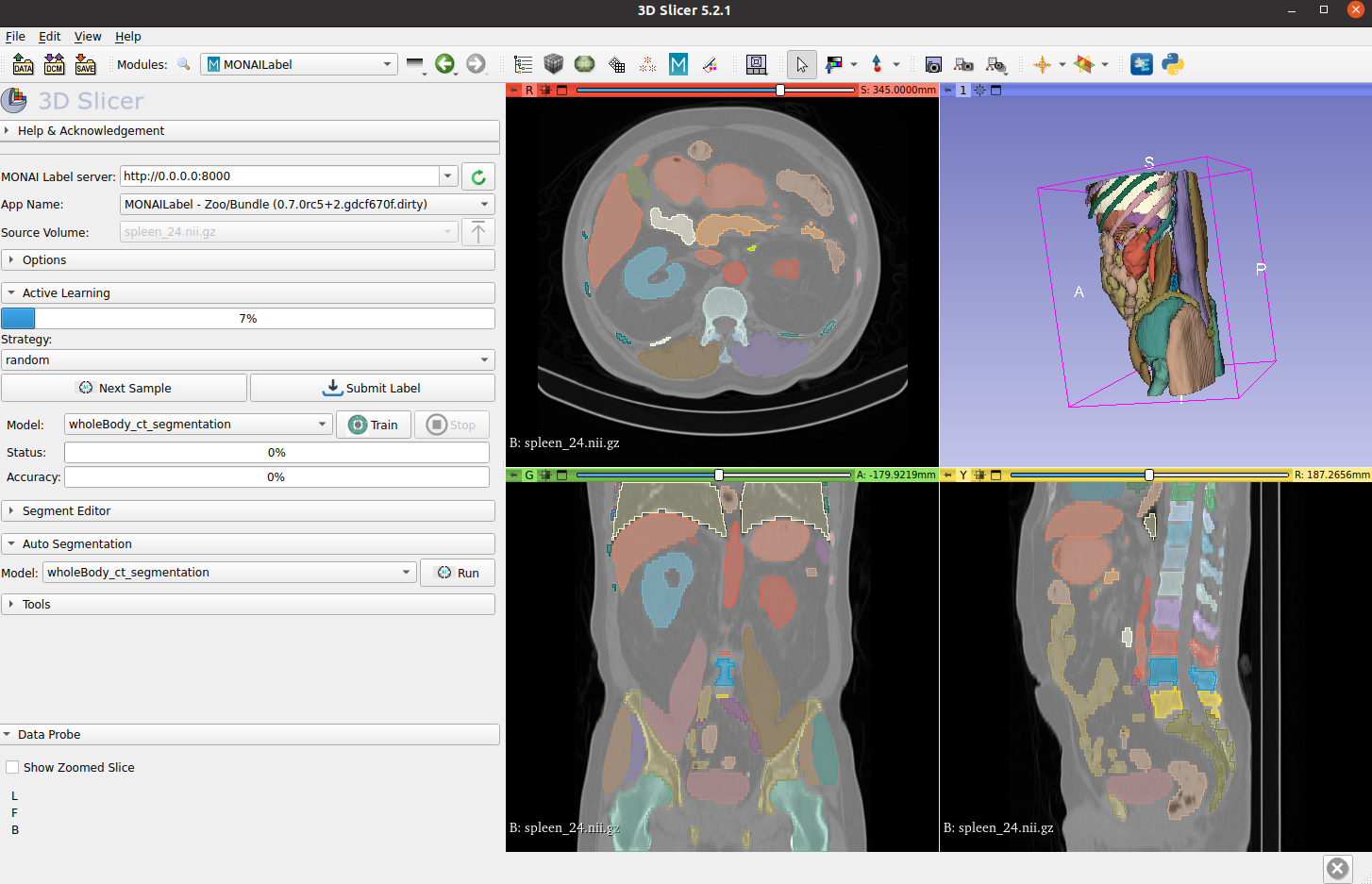
Whole-body CT segmentation
Segmenting the entirety of a whole-body CT scan from a single model is a daunting task. However, the MONAI team has risen to the challenge. They’ve developed models that segment all 104 anatomical structures from a single model:
- 27 organs
- 59 bones
- 10 muscles
- 8 vessels
Using the dataset released by the totalSegmentator team, MONAI conducted research and benchmarking to achieve fast inference times. For a high-resolution 1.5 mm model, the inference time using a single NVIDIA V100 GPU for all 104 structures is just 4.12 seconds, while the inference time using a CPU is 30.30 seconds. This is a significant improvement from the original paper’s reported inference time for a single CT scan, which took more than 1 minute.
To access the MONAI Whole Body CT Segmentation foundation model, see the MONAI Model Zoo.
For more information about the overview of all anatomical structures in whole-body CT scans, see the TotalSegmentator: robust segmentation of 104 anatomical structures in CT images whitepaper.
(Source: TotalSegmentator: robust segmentation of 104 anatomical structures in CT images)
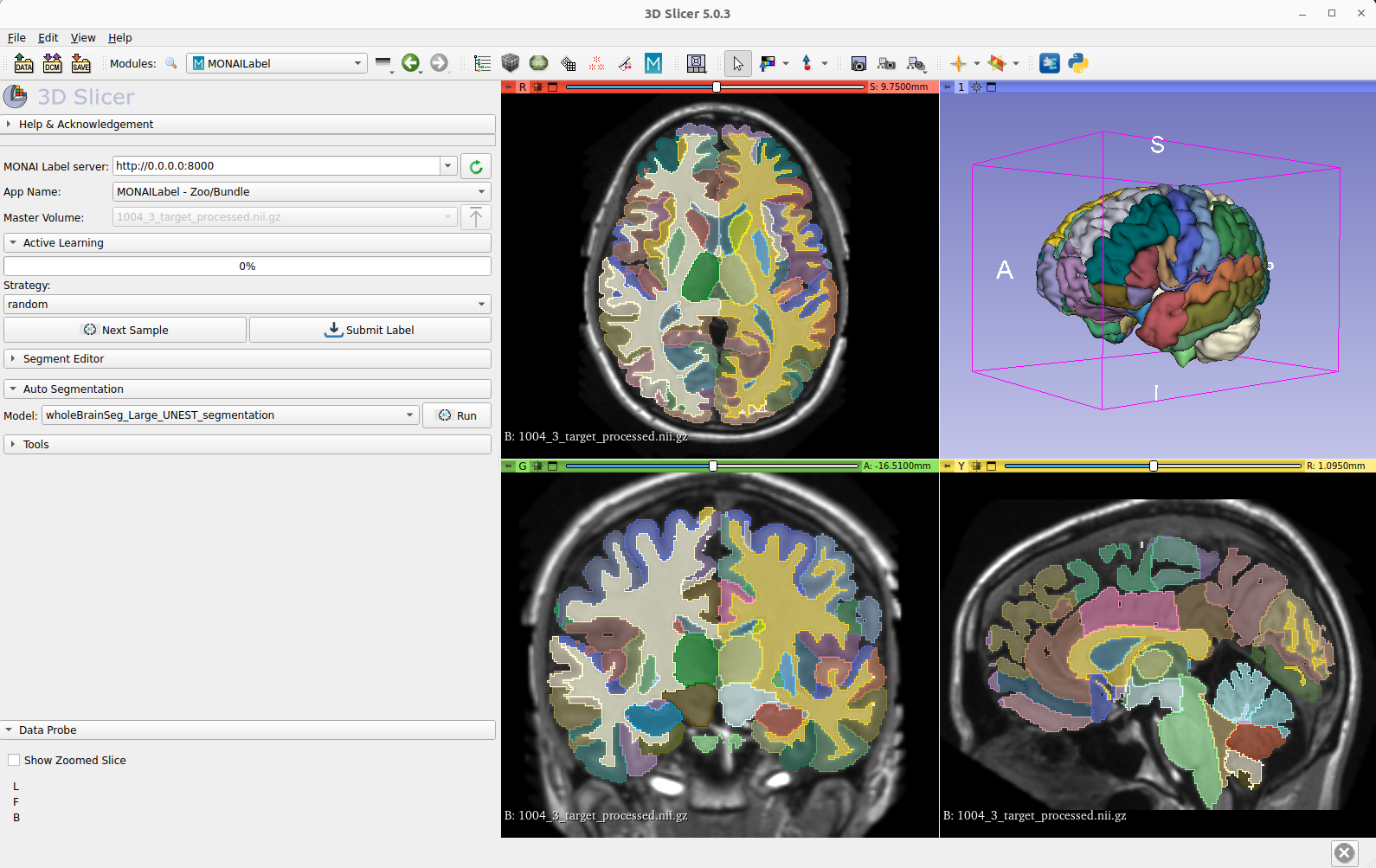
Whole-brain MRI segmentation
Whole-brain segmentation is a critical technique in medical image analysis, providing a non-invasive means of measuring brain regions from clinical structural magnetic resonance imaging (MRI). However, with over 130 substructures in human brains, segmenting anything in the brain is a difficult challenge for MRI 3D segmentation. Unfortunately, detailed annotations of the brain are scarce, making this task even more challenging for the medical imaging community.
To address this issue, the MONAI team collaborated with Vanderbilt University to develop a deep learning model that can simultaneously segment all 133 brain structures. Using 3D Slicer, the MONAI model can infer the entire brain in just 2.0 seconds. The MONAI whole brain MRI segmentation model represents a promising development in medical imaging research, offering a valuable resource for improving the accuracy of brain measurements in clinical settings.
Visit the MONAI Model Zoo to access the MONAI Whole Brain MRI Segmentation Foundation Model.
How to access medical imaging foundation models
The use of foundation models in medical image analysis has great potential to improve diagnostic accuracy and enhance patient care. However, it’s important to recognize that medical application requires strong domain knowledge.
With the ability to process large amounts of data and identify subtle patterns and anomalies, foundation models have proven to be valuable tools in the medical image analysis field. The development and refinement of these models is ongoing, with researchers and practitioners working to improve their accuracy and expand their capabilities.
Although challenges such as patient privacy and potential biases must be addressed, the use of foundation models has already demonstrated significant benefits. It is expected to play a more prominent role in healthcare in the future.
As researchers, clinicians, and users continue to focus on foundation models, the MONAI Model Zoo, a platform hosting pretrained medical image models, is amplifying its impact. Fine-tuning pretrained models is crucial to the future of medical image analysis.
The MONAI Model Zoo provides access to a diverse collection of pretrained models for various medical imaging tasks, including segmentation, classification, registration, and synthesis. By using these pre-existing models as starting points, researchers and clinicians can accelerate the development of new medical imaging applications, saving time and resources.
Join us in driving innovation and collaboration in medical imaging research by exploring the MONAI Model Zoo today.