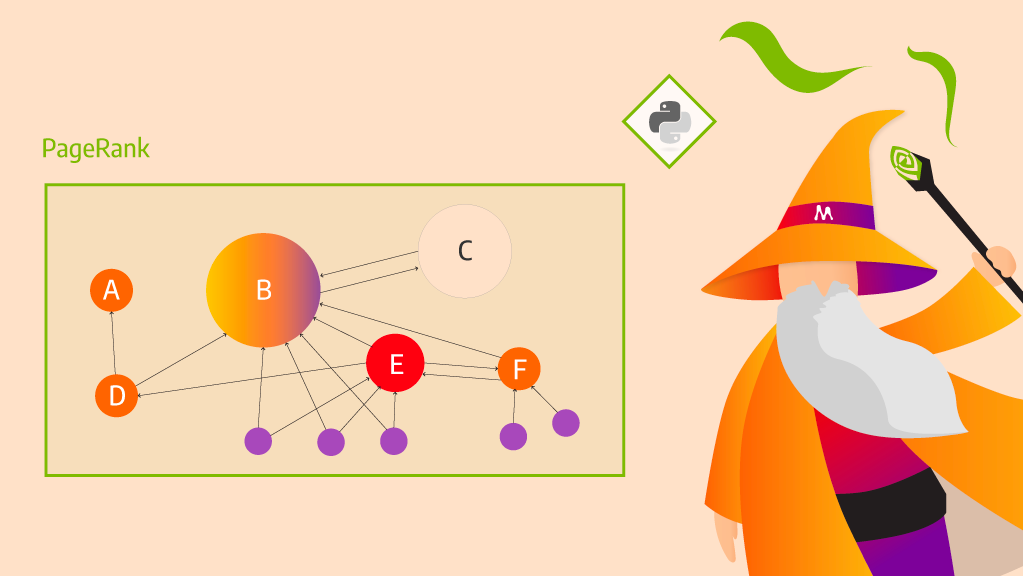
Community detection algorithms play an important role in understanding data by identifying hidden groups of related entities in networks. Social network analysis, recommendation systems, GraphRAG, genomics, and more depend on community detection. But for data scientists working in Python, the ability to efficiently analyze graph data as it grows in size and complexity can pose a problem when building responsive, scalable community detection systems.
While there are several community detection algorithms in use today, the Leiden algorithm has become a leading solution for data scientists. And for large-scale graphs in Python, this once-expensive task is now dramatically faster thanks to cuGraph and its GPU-accelerated Leiden implementation. Leiden from cuGraph delivers results up to 47x faster than comparable CPU alternatives. This performance is easily accessible in your Python workflows through the cuGraph Python library or the popular NetworkX library through the nx-cugraph backend.
This post demonstrates where the Leiden algorithm can be used and how to accelerate it for real-world data sizes using cuGraph. Read on for a brief overview of Leiden and its many applications, benchmarks of cuGraph Leiden performance against others available in Python, and an example of GPU-accelerated Leiden on larger-scale genomics data.
What is Leiden?
Leiden was developed as a modification to the popular Louvain algorithm, and like Louvain, it aims to partition a network into communities by optimizing a quality function called modularity. However, Leiden also addresses a significant drawback of Louvain: the resulting communities returned by Louvain can be poorly connected, sometimes even disconnected. By adding an intermediate refinement phase, Leiden guarantees all resulting communities are well-connected, making it a popular choice for a broad selection of applications. Leiden is quickly becoming the standard alternative to Louvain.
Where is Leiden used?
The following is just a sample of the fields that use community detection techniques such as Leiden, all of which are subject to the impact of constantly growing real-world data sizes:
- Social network analysis: Identifying communities can reveal groups of users with shared interests, facilitating targeted advertising, recommendations, and the study of information diffusion.
- Recommendation systems: Clustering users or items into communities based on their interactions allows for recommendation systems to provide more accurate and personalized suggestions.
- Fraud detection: By identifying communities of fraudulent accounts or suspicious transactions in financial networks, institutions can quickly flag and investigate fraudulent activity.
- Graph-based retrieval-augmented generation (GraphRAG): GraphRAG retrieves relevant information from a knowledge graph—a web of interconnected facts—to give an LLM better context. Leiden is often used to create categories of knowledge to assist in matching the most applicable nodes in the knowledge graph to the user’s prompt.
- Genomics: Leiden is used when analyzing single-cell genomics data for identifying groups of cells with similar gene expression profiles.
How does GPU-powered Leiden from cuGraph compare?
Several Leiden implementations available to Python developers were benchmarked using a patent citation graph consisting of 3.8 million nodes and 16.5 million edges, where the communities identified by Leiden represent related technologies. Figure 1 shows the runtime in seconds, along with the number of unique communities identified.

Software: NetworkX 3.5, cugraph/nx-cugraph 25.10; CPU: Intel Xeon Platinum 8480CL 2TB RAM; GPU: NVIDIA H100 80GB RAM
Note that because Leiden implementations use a random number generator, the communities returned are non-deterministic and vary slightly between runs. The number of communities is shown to indicate that all results are approximately equal. Most implementations, including cuGraph’s, provide parameters to tune for larger or smaller community sizes, among others. Each implementation was called with the default parameter values when possible. The source code for these benchmarks can be found in the rapidsai/cugraph GitHub repo.
As shown in Figure 1, the cuGraph GPU-accelerated Leiden implementation runs 8.8x faster than igraph’s and 47.5x faster than graspologic’s on the same citation graph. In addition to high performance, cuGraph also delivers ease of use, flexibility, and compatibility with existing Python data science workflows through multiple Python interfaces. To help you choose the right one for your project, Table 1 lists key features of each library. Leiden and many other graph algorithms are available in each.
| Speed | Ease-of-use | Dependencies | NetworkX benefits: CPU fallback, flexible graph object, popular API, hundreds of algos, graph visualization, more | Multi-GPU support | cuDF and Dask support | |
| NetworkX plus nx-cugraph | Fast | Easiest | Few | ✔ | ||
| cuGraph | Faster | Easy | More, including cuDF and Dask | ✔ | ✔ |
For detailed installation instructions, see the RAPIDS Installation guide. To get started right away with either pip or Conda, use the RAPIDS release selector.
How to use NetworkX and nx-cugraph with genomics data
Genomics datasets are massive, and growing at an explosive pace, largely due to the recent and dramatic drop in the cost of DNA sequencing. While NetworkX has an enormous following among data scientists of all fields, its pure-Python implementation means that most genomic datasets are simply too large for it, forcing scientists to learn and integrate a separate library for analytics. Fortunately, NetworkX can be GPU accelerated by enabling the nx-cugraph backend to enable data scientists to continue using NetworkX even with large data.
To demonstrate the benefit of GPU accelerated NetworkX on larger-scale genomics data, a simple example was created that reads gene expression data, builds a graph of genes with edges connecting genes based on expression correlation values, runs Leiden to identify groups of functionally related genes, and plots the communities for visual inspection. The full source code is available in the rapidsai/nx-cugraph GitHub repo. Note that the example represents a common operation in genomics—community detection using Leiden or Louvain—on actual genomics data, but is not intended to be representative of a typical genomics workflow.
The gene expression analysis data used results in a graph of 14.7K nodes and 83.8 million edges. The following code will run Leiden using nx-cugraph but will fall back to the NetworkX implementation of Louvain when nx-cugraph is not available.
Leiden is currently the only algorithm provided by nx-cugraph that does not have an alternative implementation available through NetworkX. This means that Leiden is available to NetworkX users only through nx-cugraph. For this reason, this workflow uses Louvain from NetworkX on CPU, as it provides a reasonable comparison for a user wishing to continue using NetworkX when a GPU is not present.
With nx-cugraph enabled, NetworkX identified four communities in less than 4 seconds. However, falling back to the NetworkX implementation of Louvain shows that the results are nearly identical (within tolerance of the non-determinism of both Leiden and Louvain), but performance is dramatically slower, taking nearly 21 minutes. Furthermore, since Louvain was used, the resulting communities are not guaranteed to be well-connected.
This makes NetworkX with nx-cugraph 315x faster at delivering higher quality results than NetworkX Louvain on CPU.
To run Leiden or Louvain based on the presence of the Leiden implementation (currently available only through nx-cugraph) use the following code:
%%time
try:
communities = nx.community.leiden_communities(G)
except NotImplementedError:
print("leiden not available (is the cugraph backend enabled?), using louvain.")
communities = nx.community.louvain_communities(G)
num_communities = len(communities)
print(f"Number of communities: {num_communities}")

Software: NetworkX 3.5, cugraph/nx-cugraph 25.10; CPU: Intel Xeon Gold 6128 CPU @ 3.40 GHz 48 GB RAM; GPU: NVIDIA Quadro RTX 8000 48 GB RAM
Coloring graph nodes by community and plotting is trivial in NetworkX (Figure 3).

When NetworkX does add CPU support for Leiden, either as a native Python implementation or as a separate CPU backend, users can take advantage of zero-code-change functionality by having a single “portable” function call that works, albeit possibly slower, on platforms without a GPU.
Purpose-built tools for genomics using GPU-accelerated Leiden
The previous example is intended to simply demonstrate how nx-cugraph can GPU accelerate NetworkX algorithms commonly used in genomics on real-world genomics data. To explore more realistic, purpose-built examples, check out the RAPIDS-singlecell project, which offers a library designed specifically for genomics problems.
RAPIDS-singlecell is an scverse core package based on the popular Scanpy library, supports an AnnData-compatible API, and is optimized for single-cell analysis on large datasets. The impressive speed of RAPIDS-singlecell at scale comes from cuGraph and other CUDA-X DS libraries that provide GPU acceleration for its calls to Leiden and many other algorithms. To learn more, see Driving Toward Billion-Cell Analysis and Biological Breakthroughs with RAPIDS-singlecell.
Get started running GPU-powered Leiden workflows
cuGraph provides best-in-class community detection performance through its GPU accelerated Leiden implementation, available to data scientists in Python from the cuGraph Python library or the popular and flexible NetworkX library through the nx-cugraph backend. Performance up to 47x faster, possibly more, over comparable CPU implementations mean genomics and many other applications relying on community detection can scale their data and solve bigger problems in far less time.
To get started, check out the RAPIDS Installation guide or visit the rapidsai/cugraph or rapidsai/nx-cugraph GitHub repos to run your GPU-powered Leiden workflows.