To help advance medical research while preserving data privacy and improving patient outcomes for brain tumor identification, NVIDIA researchers in collaboration with King’s College London researchers today announced the introduction of the first privacy-preserving federated learning system for medical image analysis. NVIDIA is working with King’s College London and French startup Owkin to enable federated learning for the newly established London Medical Imaging and AI Centre for Value Based Healthcare.
This new paper will be presented at MICCAI, one of the world’s top conferences on medical imaging, kicking off on Oct. 14 in Shenzhen, China. In the work, NVIDIA and King’s College London researchers describe in detail how they developed their technique.
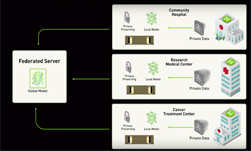
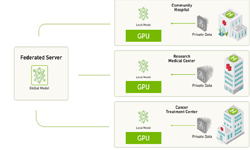
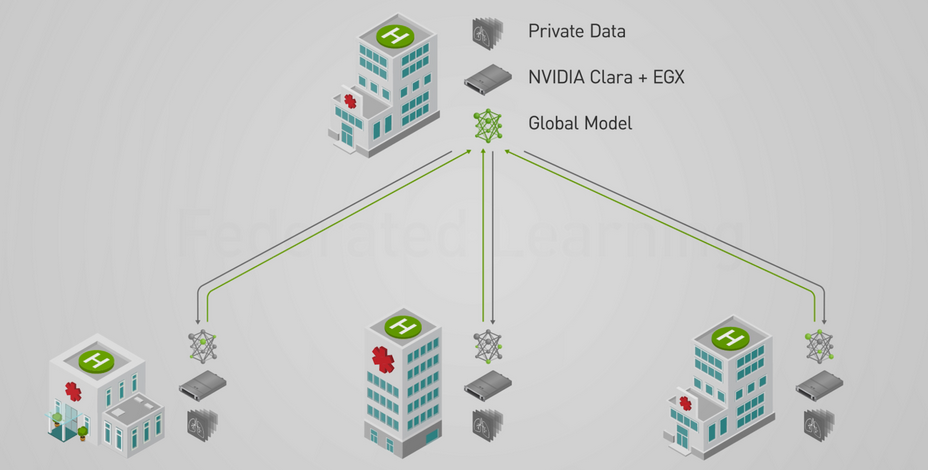
Federated learning is a learning paradigm that allows developers and organizations to train a centralized deep neural network (DNN) with training data distributed across multiple locations. This makes it possible for organizations to collaborate on a shared model, without needing to directly share any clinical data.
“Federated learning allows collaborative and decentralized training of neural networks without sharing the patient data,” the researchers stated in their paper. “Each node trains its own local model and, periodically, submits it to a parameter server. The server accumulates and aggregates the individual contributions to yield a global model, which is then shared with all nodes.”
Although federated learning can provide high security in terms of privacy, there are still ways to reconstruct data by model inversion, the researchers explained. To help make federated learning even safer, the researchers investigate the feasibility of using the ε-differential privacy framework, a way to formally define privacy loss, to protect patient and institutional data with a strong privacy guarantee. To ensure patient privacy is priority, differential privacy and other state-of-the-art privacy protection techniques are being built into the Owkin architecture.
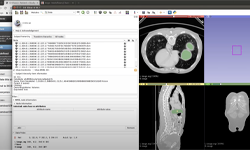
The experiments for this breakthrough were performed on brain tumor segmentation data from the BraTS 2018 dataset. The BraTS 2018 dataset contains MRI scans of 285 patients with brain tumors.
The dataset is used here for evaluating federated learning algorithms on the multi-modal and multi-class segmentation task. On the client-side, the team adapted a state-of-the-art training pipeline originally designed for data-centralized training and implemented it as part of the NVIDIA Clara Train SDK.
For training and inference, the team used NVIDIA V100 Tensor Core GPUs.
When comparing federated learning to a data-centralized system, the proposed approach can achieve a comparable segmentation performance without sharing institutional data.
Moreover, the experimental results show a natural tradeoff between privacy protection and the quality of the trained model. Still, with the sparse vector technique, the federated learning system can provide rigorous privacy protection with only a reasonably small cost in model performances.
Deep learning is a powerful technique for automatically extracting knowledge from medical data, the NVIDIA team explained. Federated learning has the potential of effectively aggregating knowledge across institutions learned locally from private data, thus further improving the accuracy, robustness, and generalization ability of the deep models, they added.
“This research is an important step towards the deployment of secure federated learning, which will enable data-driven precision medicine at large scale,” the NVIDIA researchers stated.
Read more about the paper in this tutorial: https://www.nvidia.com/en-us/events/miccai/
Editor’s note: Since this post was first published we have updated the post to reflect Owkin’s contribution to this work.