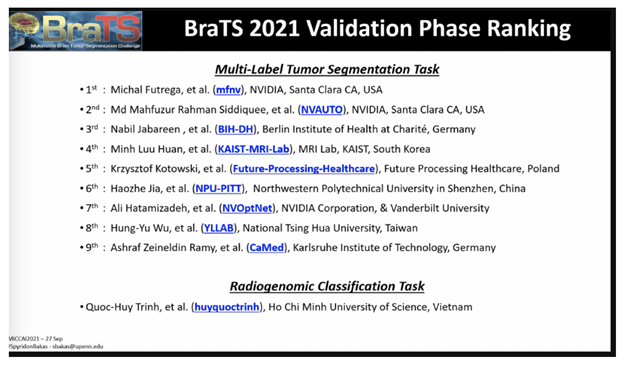
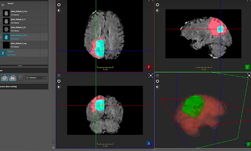
NVIDIA data scientists this week took three of the top 10 spots in a brain tumor segmentation challenge validation phase at the prestigious MICCAI 2021 medical imaging conference.
Now in its tenth year, the BraTS challenge tasked applicants with submitting state-of-the-art AI models for segmenting heterogeneous brain glioblastomas sub-regions in multi-parametric magnetic resonance imaging (mpMRI) studies, which is an extremely challenging task.
Participants could also focus on a second task of classification methods to predict the MGMT promoter methylation status.
More than 2,000 AI models were submitted to the challenge, which is jointly organized by the Medical Image Computing and Computer Assisted Interventions society, the Radiological Society of North America and the American Society of Neuroradiology.
NVIDIA developers ranked No. 1, No. 2 and No. 7 in the challenge’s validation phase, each creating different types of AI model approaches for tumor segmentation — including an optimized U-Net model, a SegResNet model with automatic hyperparameter optimization, and a Swin UNETR model which is a transformer-based approach for computer vision.
The NVIDIA winners all utilized the open-source PyTorch framework MONAI (Medical Open Network for AI), which is a freely available, community-supported initiative built by academic and industry leaders to standardize best practices for deep learning in healthcare imaging.
Optimized U-Net for Brain Tumor Segmentation – Rank #1
This optimizedU-Net model, which placed first in the BraTS validation phase, is an encoder-decoder type of convolutional network architecture for fast and precise segmentation of images. It achieved a normalized statistical ranking score of 0.267.
The starting point for designing Optimized U-Net was the BraTS 2020 winning solution: nnU-Net for Brain Tumor Segmentation. The team’s goal was to optimize the U-Net architecture as well as the training schedule. To find the optimal model architecture, the data scientists ran an extensive ablation study and found that vanilla U-Net with deep supervision yielded the best results.
The U-Net model was further optimized by adding an additional channel to the input with one-hot encoding for foreground voxels, increasing both encoder depth by one level and number of convolution channels. The model was trained better and faster with deep supervision by adding two additional output heads on lower decoder levels which allowed for better gradient flow and more accurate predictions. MONAI was used for data preprocessing to clean and denoise the data, as well as for inferencing of the model. Data augmentation, a technique that is used to artificially enlarge the size of the dataset, was used and powered by the NVIDIA Data Loading Library (DALI), which addresses the problem of the CPU bottleneck by offloading data augmentations to the GPU.
NVIDIA PyTorch containers with the latest PyTorch, cuDNN and CUDA versions were used to optimize the U-Net model for fast training. Automatic Mixed Precision (AMP) was used to reduce the memory footprint of the AI model by a factor of 2 and speed up training. Training was done on eight NVIDIA A100 GPUs for 1000 epochs, which allows for 2x speed up versus V100 GPUs. Nearly 100% of the GPUs were used, demonstrating the optimization of the network to use GPUs efficiently. This 3D U-Net model can be used for any 3D modality, such as MRI and CT. To learn more GPU efficient nnU-Net implementation.
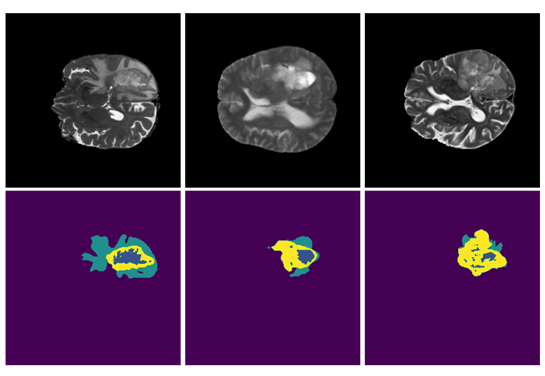
SegResNet: Redundancy Reduction in Semantic Segmentation of 3D Brain MRIs – Rank #2
This approach, which ranked second in the BraTS challenge and will be made available in MONAI, is based on MONAI components and aims to demonstrate the utility and flexibility of its applications. The main model is the SegResNet architecture from MONAI, a standard encoder-decoder based convolutional neural network (CNN) similar to U-Net. The approach is a part of automation (AutoML) initiative for MONAI, with the hyperparameters automatically selected using hyperparameters optimization and tuning.
The method achieved top performance in the validation stage leaderboard (team NVAUTO), and achieved a rank of 0.272 in the combined ranking, based on per case ranking and perturbation analysis. The organizers indicated that this rank is not statistically significantly different from the 1st rank solution, and both methods are considered to be statistically similar.
Two new contributions were added in this work to further boost the performance. First, the training procedure was modified to enforce certain properties on the learned feature representations. By borrowing ideas from self-supervised literature, the feature dimension was regularized to have minimal redundancy between different anatomical regions. At the same time, regions of the same anatomy were encouraged to be similar. This allows for better network behavior and generalization. Second, an adaptive ensembling technique was used to adaptively select a subset of models for ensembling. This helped to avoid potential outliers in some of the models predictions, and further boost the final ensemble performance.
The method was implemented in MONAI, based on PyTorch, and trained on four NVIDIA V100 GPUs for 300 epochs using a Dice loss function for 16 hours. The team used NVIDIA-provided PyTorch containers and AMP to achieve fast training during hyperparameters optimization. The method was trained with 5-fold cross validation, and best performing checkpoints were kept from each fold. Overall, 25 model checkpoints were saved, but using the adaptive ensembling, only a half of them were used in the final prediction. Since the approach is fully CNN-based, the inference time is fast and can be done in one step on the whole input image, without any sliding windows. It takes less than a second for a single model inference, which allows for high throughput and nearly real-time results, which can be important in clinical settings.
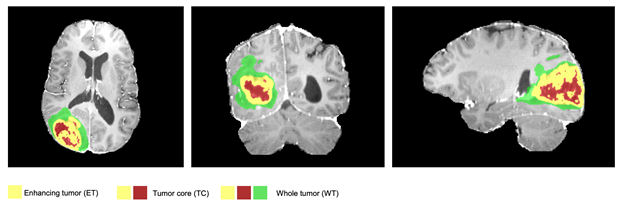
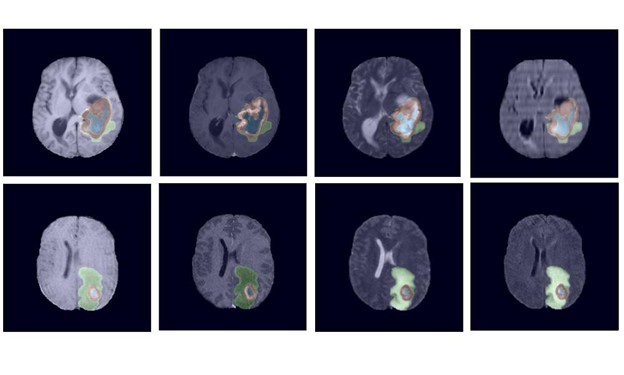
visible labels (a union of green, yellow and red labels), the tumor core (TC) class is a
union of red and yellow, and the enhancing tumor core (ET) class is shown in yellow
(a hyperactive tumor part).
Swin UNETR: Shifted Window Transformers for 3D Semantic Segmentation of Brain Tumors – Rank #7
Swin UNETR, which ranked seventh in the BraTS challenge, is a transformer-based model instead of a CNN model. Implemented in MONAI, it achieved an average Dice score of 92.94% and Hausdorff distance of 1.7 across whole tumor, tumor core and enhanced tumor segmentation classes.
Transformers are a new class of deep learning-based models that are used in sequence-to-sequence prediction tasks. In their original formulation, they are composed of an encoder and decoder. The encoder consists of layers with multi-headed self-attention followed by multi-layer perceptrons. The inputs to each of these modules are added to the output via residual blocks and normalized. The self-attention layers learn the weighted sum of values calculated from hidden layers and can highlight the important features of a given input sequence. Although they were initially proposed for machine translation tasks in natural language processing, they have since been successfully applied to other fields such as computer vision and protein drug generation and achieved state-of-the-art performance in various benchmarks.In computer vision, transformers have achieved new state-of-the-art performance in various benchmarks. Swin UNETR is a new way of building a model utilizing the continued advances in GPU architecture and performance.
Swin transformers are hierarchical transformers whose representation is computed with Shifted WINdows (Swin). These transformers are well suited for computer vision tasks such as object detection, image classification, semantic segmentation, and more. Swin transformers can model the differences between two domains such as variations in the scale of objects and the high resolution of pixels in the images more efficiently and can serve as a general-purpose pipeline for vision. This NVIDIA Swin UNETR model leverages a Swin transformer encoder to directly utilize 3D patches of input data without relying on CNNs for feature extraction. This allows the Swin UNETR to have access to contextual multi-modal information in the input data and efficiently process them as tokenized embeddings into the transformer encoder. The transformer-based encoder of Swin UNETR is then connected to a CNN decoder in a U-shape architecture via skip connections to make the final segmentation predictions.
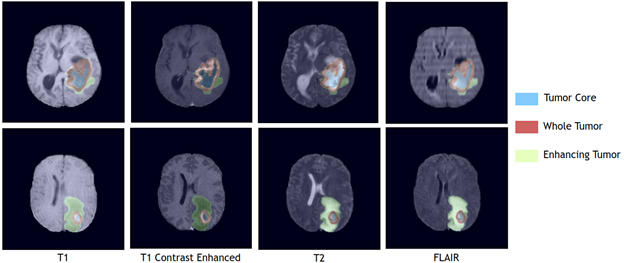
NVIDIA’s Swin UNETR model was trained on a NVIDIA DGX-1 cluster using eight GPUs with an initial learning rate of 0.0008 and using the AdamW optimization algorithm. Random patches of 128*128*128 input data with data augmentation strategies of random axis mirror flip and intensity shift were used. Each training round takes 24 hours to complete. In comparison to commonly used CNN-based segmentation models, Swin UNETR is more efficient in terms of number of FLOPs and has a moderate model-complexity in terms of the number of trainable parameters. It can be trained and used for inference in an efficient manner. For model optimization, a generic soft Dice loss function was used for learning to segment various brain tumor regions with a seperate output channel dedicated to each class. The model was trained using a 5-fold cross validation scheme on the entire BRATS21 training set wherein the final segmentation output is computed by averaging the outputs of 10 models from 2 different 5-fold cross validations.
Find the full leaderboard of BraTS participants here.