The world of healthcare is under a giant spotlight these days. In these unprecedented times, everyone is focusing on keeping loved ones safe and contributing to the community as much as possible. It is amazing to see how the community is uniting in the past two months, which has led to a global, rapid emergence of SARS-CoV-2 projects across the life sciences.
At NVIDIA, we realize the importance of working with the community to overcome healthcare challenges. To accelerate genomics research for COVID-19, we announced free access to Clara Parabricks for any COVID-19 related research. The response has been remarkable, with over 600 requests from over 88 countries.
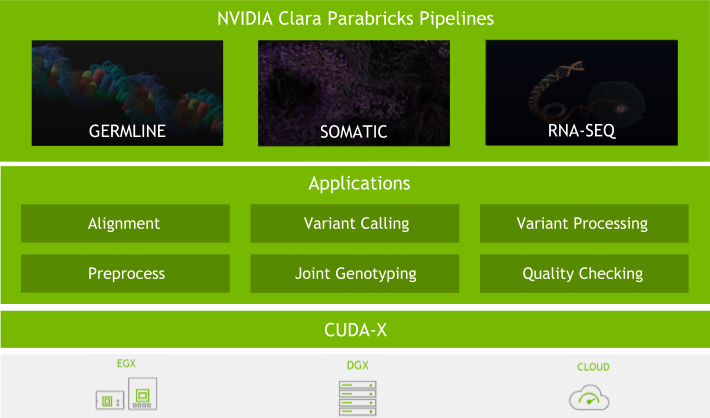
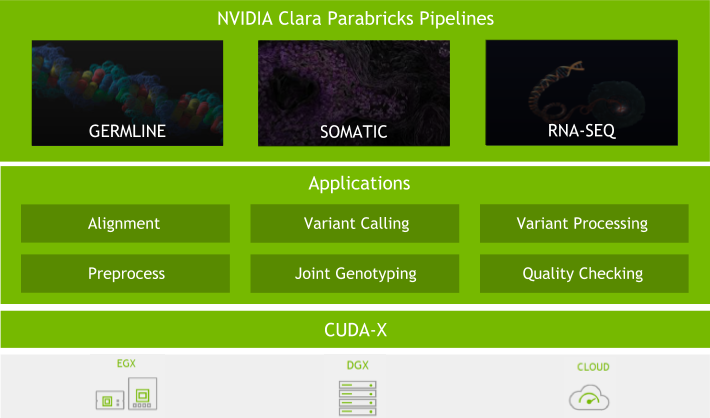
Because of this, we have put significant effort into the software. With every major Clara Parabricks Pipelines update, we strive to provide you with new tools and features, better accuracy, and more stability to analyse next-generation sequencing (NGS) data. Here are some of the exciting updates to the Clara Parabricks Pipelines software in version 3.0.
Thirty percent faster
The team has worked hard to change the inner workings of GPU BWA-Mem, coordinate sorting, mark-duplicates, and accelerated GPU haplotypecaller to provide more than 30% more acceleration on existing hardware that has Volta architecture or later.
As an example, a germline analysis of a 30x whole genome human sequencing data can now be finished in 30 minutes with an 8-V100 server. Similar speedups should be expected on compatible servers with T4, Titan, and RTX cards as well. This means throughput at your datacenter or cluster just increased by 30%, with a comparable reduction in the per-genome cost of analysis.
New RNA analysis
Many users wanted accelerated RNA-analysis pipelines, due to the recent explosion in RNA-Seq data. With this release, we accelerated the STAR aligner (both single and two-pass) and STAR-Fusion. Performance improvements may vary depending on dataset and analysis, but initial acceleration is about 10x when compared to the baseline CPU version.
This is an early release version, and we will continuously improve the RNA-accelerated tools in upcoming months, just as we demonstrated with the germline pipeline.
GATK 4.1 and DeepVariant 0.10.0 support
Parabricks GPU-accelerated tools have been ported to be compatible with GATK4.1. As with previous versions, we have 100% equivalent BAM output and 99.99% concordance with the CPU GATK 4.1. We will continue to support all major updates of GATK when available.
The DeepVariant team recently released DeepVariant 0.10.0, which was exciting because of their inclusion of PacBio models. Parabricks supports the entire PacBio DeepVariant workflow and continues to provide the acceleration and throughput benefits.
DNAnexus availability
Clara Parabricks Pipelines are now available in the DNAnexus App Library, making it easy for you to run the analysis on their platform. For more information, see DNAnexus Partners with NVIDIA Clara Parabricks to Accelerate NGS Data Processing.
Stay tuned
We have added support for multiple new features and options in existing tools. We also notify you about suboptimal server configuration at runtime for improved performance debugging. Several bug fixes have been completed and error messages have been updated.
For more information, see Clara Parabricks Pipelines.