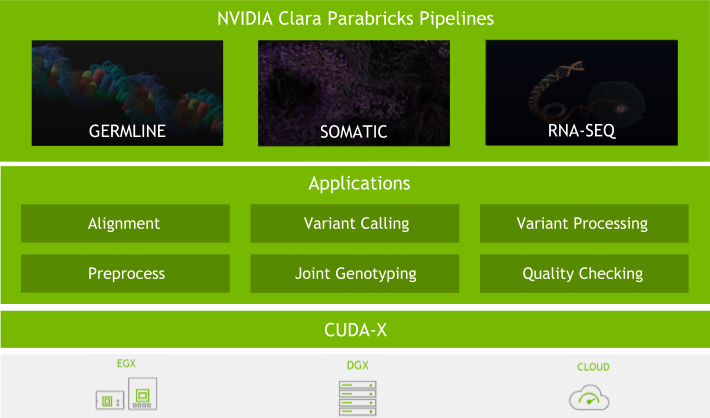
Today, NVIDIA is releasing the Clara Genomics Analysis SDK – an open source toolkit for biological sequence analysis, that is part of Clara Genomics.
The last few years have seen a revolution in genome sequencing. New high-throughput sequencing techniques such as those developed by Oxford Nanopore, PacBio and Illumina have led to increases in throughput and quality as well as large reductions in the cost of sequencing. Large-scale genomics projects such as the 100,000 Genomes Project and Darwin Tree of Life Project are making genomic datasets of unprecedented size available to researchers who want to understand the genetic basis of disease and evolution.
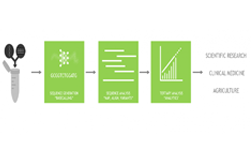
Along with large-scale collection of genomic data, the scientific community also needs computational methods for its analysis. We believe that GPUs have an important role to play in this and are developing Clara Genomics Analysis to enable researchers to study and understand ever larger and more complex genomic datasets.
Why GPUs?
GPUs have thousands of cores and are optimised for high-throughput, highly parallel applications such as those commonly found in genomic analysis. GPUs are in fact already used extensively in genomics, enabling GPU-accelerated GATK, and Oxford Nanopore’s basecallers. Kipoi model zoo contains a number of GPU-enabled, ready-to-use trained models for genomics. Our aim for Clara Genomics Analysis is to provide a platform for scientific researchers to enable faster analysis of their data and for scientific software developers to easily GPU accelerate their applications.
“GPUs are poised to play a key role in our efforts to dramatically speed up crop genome assembly and some of the downstream processes like whole genome comparison and pan genome analyses. We therefore happily welcome the open source release of the NVIDIA Clara Genomics SDK,” said Antoine Janssen, Team Lead for Genome Bioinformatics at Keygene.
Example use case: GPU acceleration of Racon
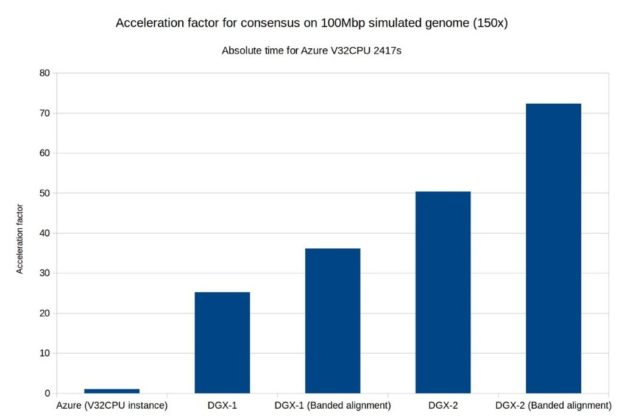
As part of Clara Genomics Analysis we provide the cudapoa module. This module is a GPU-accelerated implementation of the Partial Order Alignment (POA) algorithm for multiple-sequence alignment. Cudapoa is heavily influenced by the SPOA package and in many cases can be considered a GPU-accelerated replacement for it. Using cudapoa, we GPU-enabled Racon, a consensus module for raw de novo genome assembly of long uncorrected reads. We also investigated a new alignment algorithm (banded alignment) for the POA algorithm and were able to achieve significant speedups.
Future work
This preview release is only the beginning. Our roadmap includes a Python API as well as GPU-enabled DNA read overlapping and alignment. We are always looking for feedback and collaborations.