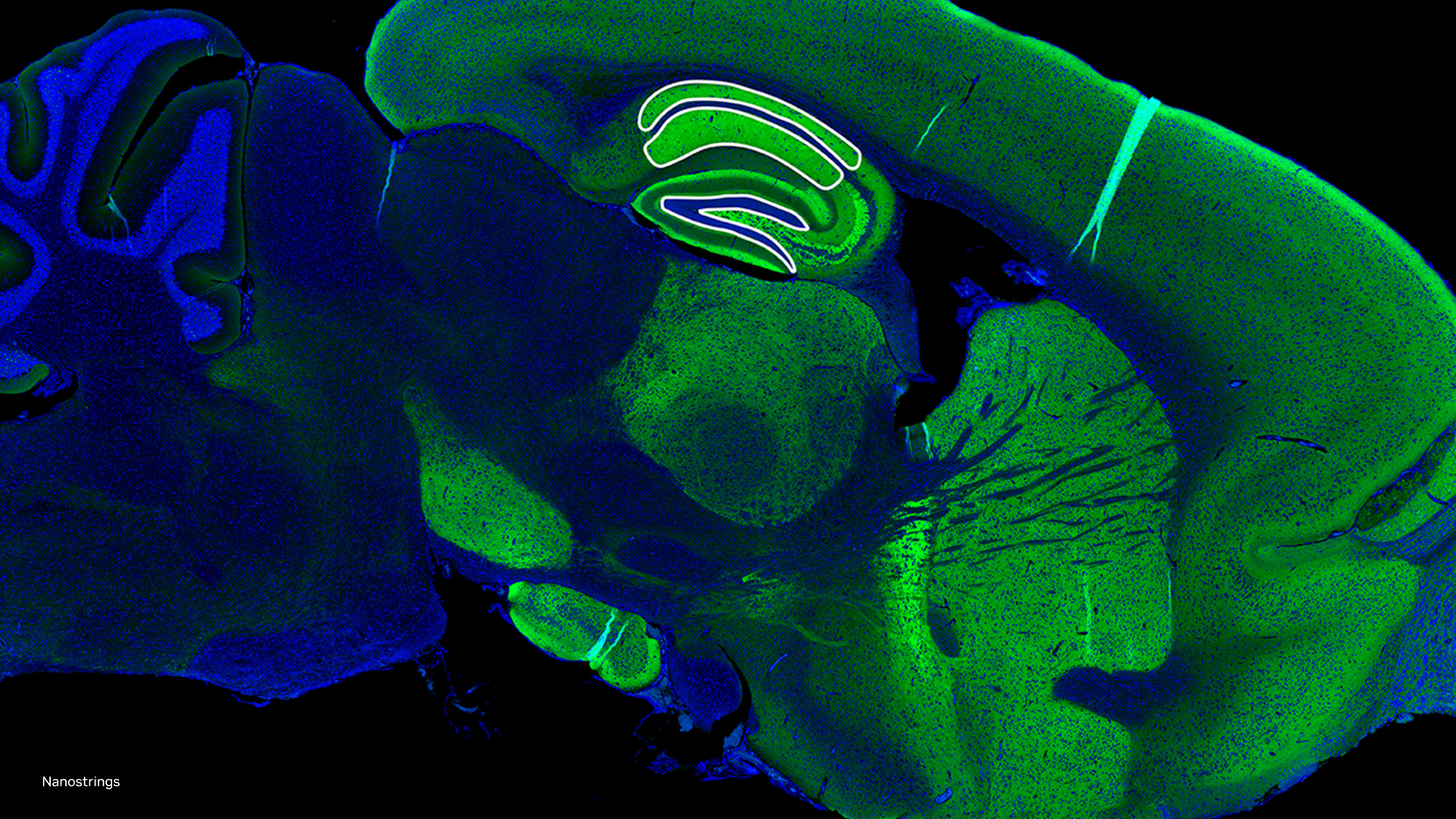
University of Alberta scientists developed a deep learning-based prostate cancer diagnostic platform that only uses a single drop of blood which will allow men to bypass the current painful biopsy methods.
Using a GTX 1060 GPU, CUDA and the MathWorks Neural Network Toolbox, the scientists’ trained their model on information from millions of cancer cell nanoparticles in the blood to recognize the unique fingerprint of aggressive prostate cancer.
To test their method, they evaluated a group of 377 men who were referred to their urologist with suspected prostate cancer and found that their system called Extracellular Vesicle Fingerprint Predictive Score (EV-FPS) correctly identified men with aggressive prostate cancer 40 percent more accurately than the most common test in wide use today.

“Higher sensitivity means that our test will miss fewer aggressive cancers,” said John Lewis, the Alberta Cancer Foundation’s Frank and Carla Sojonky Chair of Prostate Cancer Research at the University of Alberta. “For this kind of test you want the sensitivity to be as high as possible because you don’t want to miss a single cancer that should be treated.”
The team says their EV-FPS test can eventually eliminate up to 600,000 unnecessary biopsies, 24,000 hospitalizations and up to 50 percent of unnecessary treatments for prostate cancer each year in North America alone. They plan to bring the test to market through their university spin-off company Nanostics Inc.
Read more >
Related resources
- DLI course: Image Classification with TensorFlow: Radiomics?€?1p19q Chromosome Status Classification
- GTC session: Early Diagnosis of Cancer Cachexia Using Body Composition Index as the Radiographic Biomarker
- GTC session: How Artificial Intelligence is Powering the Future of Biomedicine
- GTC session: Accelerate your Workflows and Gain Competitive Advantage With AI Workstations (Presented by Z by HP)
- NGC Containers: MATLAB
- Webinar: Isaac Developer Meetup #2 - Build AI-Powered Robots with NVIDIA Isaac Replicator and NVIDIA TAO