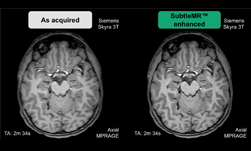
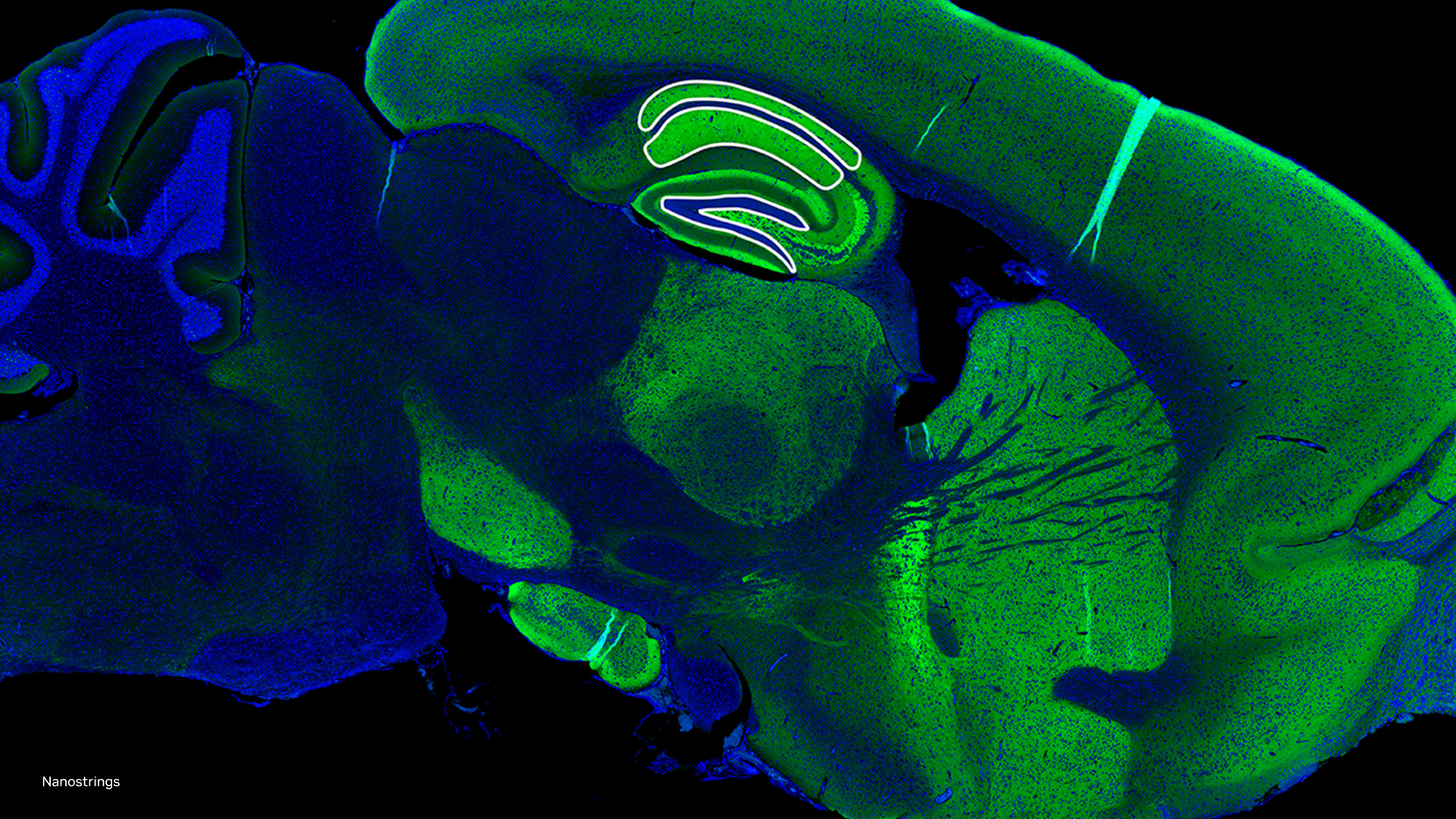
To help accelerate microscopic systems, researchers from the Salk Institute, the University of Texas at Austin, fast.AI, and others, developed a new AI-based microscopy approach that has the potential to make microscopic techniques used for brain imaging 16 times faster.
“Point scanning imaging systems are perhaps the most widely used tools for high-resolution cellular and tissue imaging,” the researchers stated in their paper Deep Learning-Based Point-Scanning Super-Resolution Imaging published on bioRxiv. “Like all other imaging modalities, the resolution, speed, sample preservation, and signal-to-noise ratio of point scanning systems are difficult to optimize simultaneously.”
To tackle this challenge, Uri Manor at the Salk Institute started with an image processing technique called deconvolution. The approach has been used by astronomers to attain greater resolution in telescope images of stars and planets.
After the team gathered enough data to build a training dataset, which consisted of high resolution and super-resolution images, the team trained their models using a ResNet-based U-Net convolutional neural network.
“To train a model for this purpose, many perfectly aligned high- and low-resolution image pairs are required. Instead of manually acquiring high- and low-resolution image pairs for training, we opted to generate semi-synthetic training data by computationally “crappifying” high-resolution images to simulate what their low-resolution counterparts might look like when acquired at the microscope,” the researchers explained in their paper.
Using NVIDIA GPUs at the Maverick Supercomputer at the Texas Advanced Computing Center, as well as NVIDIA TITAN RTX GPUs, and V100s on separate workstations, the team generated their synthetic data and trained their models.
Final models were generated using the fast.ai deep learning library, along with the cuDNN-accelerated PyTorch deep learning framework and NVIDIA Quadro P6000 GPUs. The GPUs were used for both training and inference.
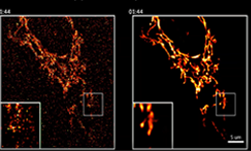
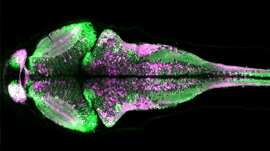
Side by side versions of mitochondria live imaging with and without denoising filters. [Credit: Salk Institute]
After the models were trained, the team put their system to the test, applying it to images created in other labs with different microscopes.
“Usually in deep learning, you have to retrain and fine-tune the model for different data sets,” Manor told the Texas Advanced Computing Center’s blog. “But we were delighted that our system worked so well for a wide range of sample and image sets.”
Manor and team hope to develop software that does reconstruction in real-time, so that researchers can see super-resolution images right away.
“Remarkably, our EM PSSR model could restore undersampled images acquired with different optics, detectors, samples, or sample preparation methods in other labs,” the team explained in their paper. “We show that undersampled confocal images combined with a multi-frame PSSR model trained on Airyscan time-lapses facilitate Airyscan-equivalent spatial resolution and SNR with ~100x lower laser dose and 16x higher frame rates than corresponding high-resolution acquisitions. In conclusion, PSSR facilitates point-scanning image acquisition with an otherwise unattainable resolution, speed, and sensitivity.”
The team has published their corresponding code on GitHub.