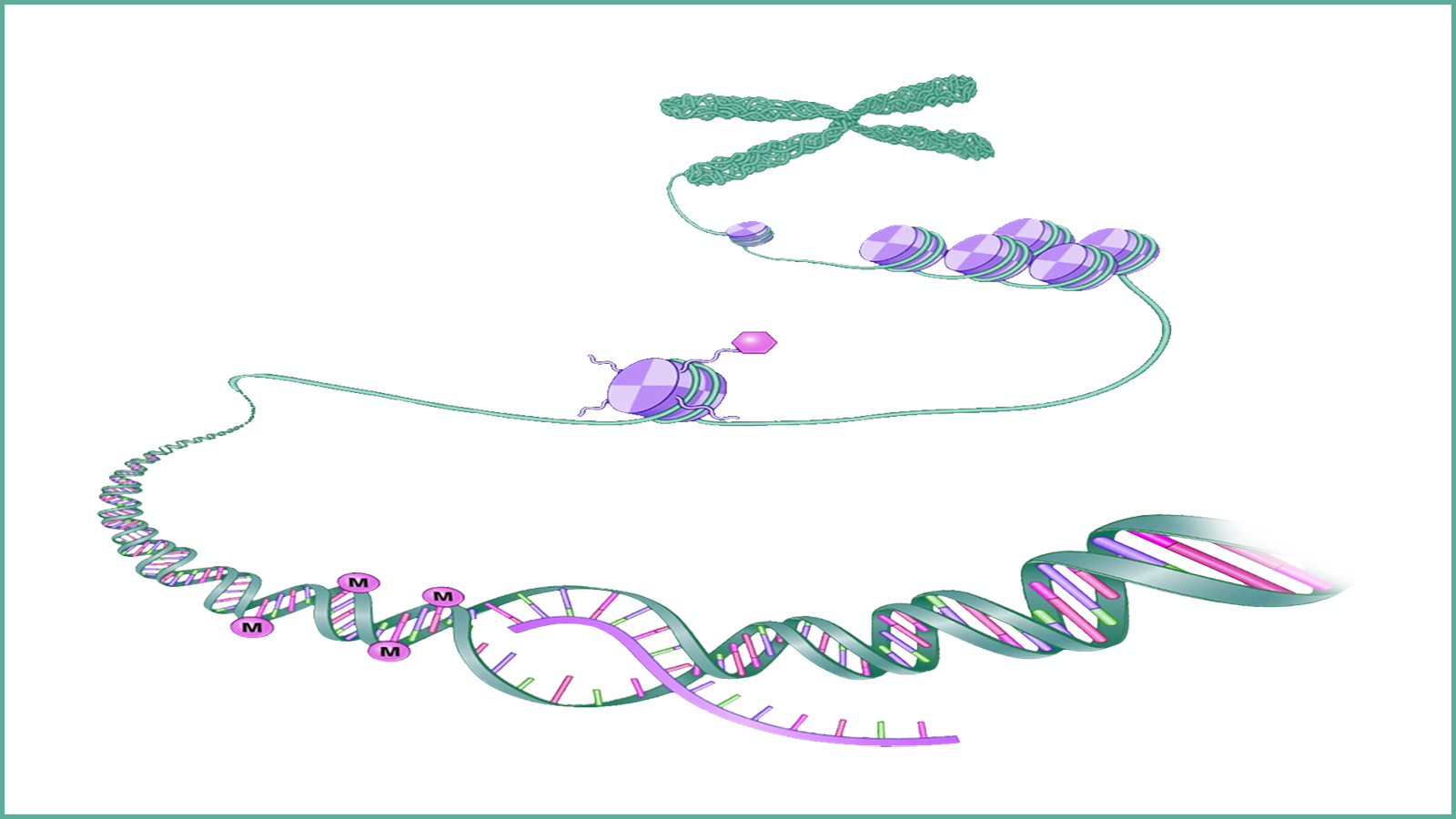
A collaboration between InstaDeep, the Technical University of Munich (TUM), and NVIDIA has led to the development of multiple super-computing scale foundation models for genomics. These models demonstrate state-of-the-art performance across many prediction tasks, such as promoter and enhancer site predictions.
The joint team of researchers showed that large language models (LLMs) trained on genomics can generalize across a plethora of genomic tasks. Previous approaches required specialized models. A sneak peek at the results will be presented at the upcoming JP Morgan Healthcare Conference, during NVIDIA Healthcare VP Kimberly Powell’s invited talk on January 12.
The team used Cambridge-1, a supercomputer launched by NVIDIA, to train a variety of large language models (LLMs), from 500M to 2.5B parameters. The models were trained on a diverse collection of genomic datasets to explore the role of model scale and data diversity on downstream task performance.
Classification tasks included the prediction of enhancer and promoter sequences and transcription factor binding sites. These tasks can help with understanding the dynamics of how DNA is translated into RNA and proteins, unlocking new clinical applications.
For each of the identified tasks in the study, the performance increased monotonically with model scale and dataset diversity. Compared to specialized state-of-the-art model baselines, the largest 2.5B parameter LLM trained on a multi-species dataset achieved equivalent or superior performance in 15 out of 18 tasks.
These results were achieved using parameter-efficient fine-tuning. Relying on pretrained embeddings extracted from various layers of the transformer model, together with a simple shallow perception (MLP) or logistic regression, was enough to achieve equivalent or superior performance in 11 tasks.
Applying this probing strategy across all layers of each model checkpoint and each task resulted in 1.2M MLP models trained. The study provided a detailed analysis of various aspects of training and using LLMs, such as the role of different layers on downstream task performance.
Direct comparisons of sequence diversity at a fixed model scale showed important gains, as did increasing the model scale. For example, the 500M parameter model trained on only the human reference genome performed less well than the same model trained on the 1000 Genomes dataset.
Similarly, the 2.5B parameter model trained on the 1000 Genomes dataset performed better than any 500M parameter model. It did not perform as well as the same model trained on a custom multi-species dataset, even when the downstream performance was measured on tasks concerning only the human genome.
The researchers observed that not all embeddings were created equally. While common wisdom suggests using the last layer of the LLM for downstream predictions, it was surprising that intermediate layers produced representations with markedly higher performance on downstream tasks.
“We believe these are the first results that clearly demonstrate the feasibility of developing foundation models in genomics that truly generalize across tasks,” said Karim Beguir, InstaDeep’s CEO. He added, “In many ways, these results mirror what we have seen in the development of adaptable foundation models in natural language processing over the last few years, and it’s incredibly exciting to see this now applied to such challenging problems in drug discovery and human health.”
Cambridge-1 was critical to the success of the project, which needed high-performance computing infrastructure to train such large models with the receptive field required to capture long-range interactions in the genome.
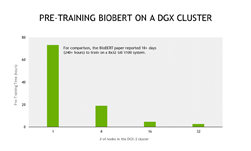
The researchers experimented with a variety of approaches, including multiple attention mechanisms, model scales, and tokenizer schemes. They finally achieved the best-published performance across tasks using a 2.5B parameter sparse attention model trained across 16 NVIDIA DGX A100 nodes (128 A100 80GB GPUs).
In future work, the team plan to explore further downstream task performance improvements by fine-tuning the models directly and will continue their collaboration on architectural innovations for large language models applied to genomics. InstaDeep was one of the first NVIDIA inception members to get access to Cambridge-1.