Image data can generally be described through two dimensions (rows and columns), with a possible additional dimension for the colors red, green, blue (RGB). However, sometimes further dimensions are required for more accurate and detailed image analysis in specific applications and domains.
For example, you may want to study a three-dimensional (3D) volume, measuring the distance between two parts or modeling how that 3D volume changes over time (the fourth dimension). In these instances, you need more than two dimensions to make sense of what you are seeing.
Greater image understanding requires enhanced capability
Multidimensional image processing, or n-dimensional image processing, is the broad term for analyzing, extracting, and enhancing useful information from image data with two or more dimensions. It is particularly helpful and needed for medical imaging, remote sensing, material science, and microscopy applications.
Some methods in these applications may involve data from more channels than traditional grayscale, RGB, or red, green, blue, alpha (RGBA) images. N-dimensional image processing helps you study and make informed decisions using devices enabled with identification, filtering, and segmentation capabilities.
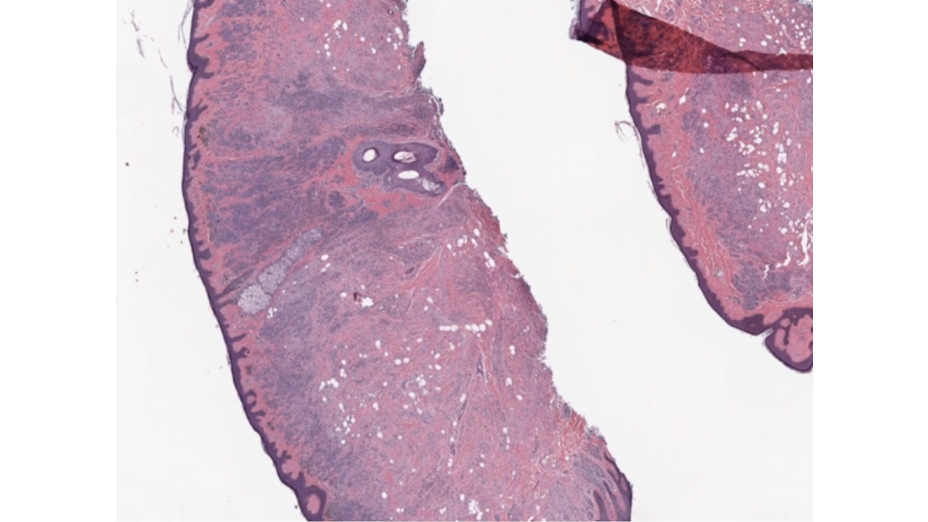
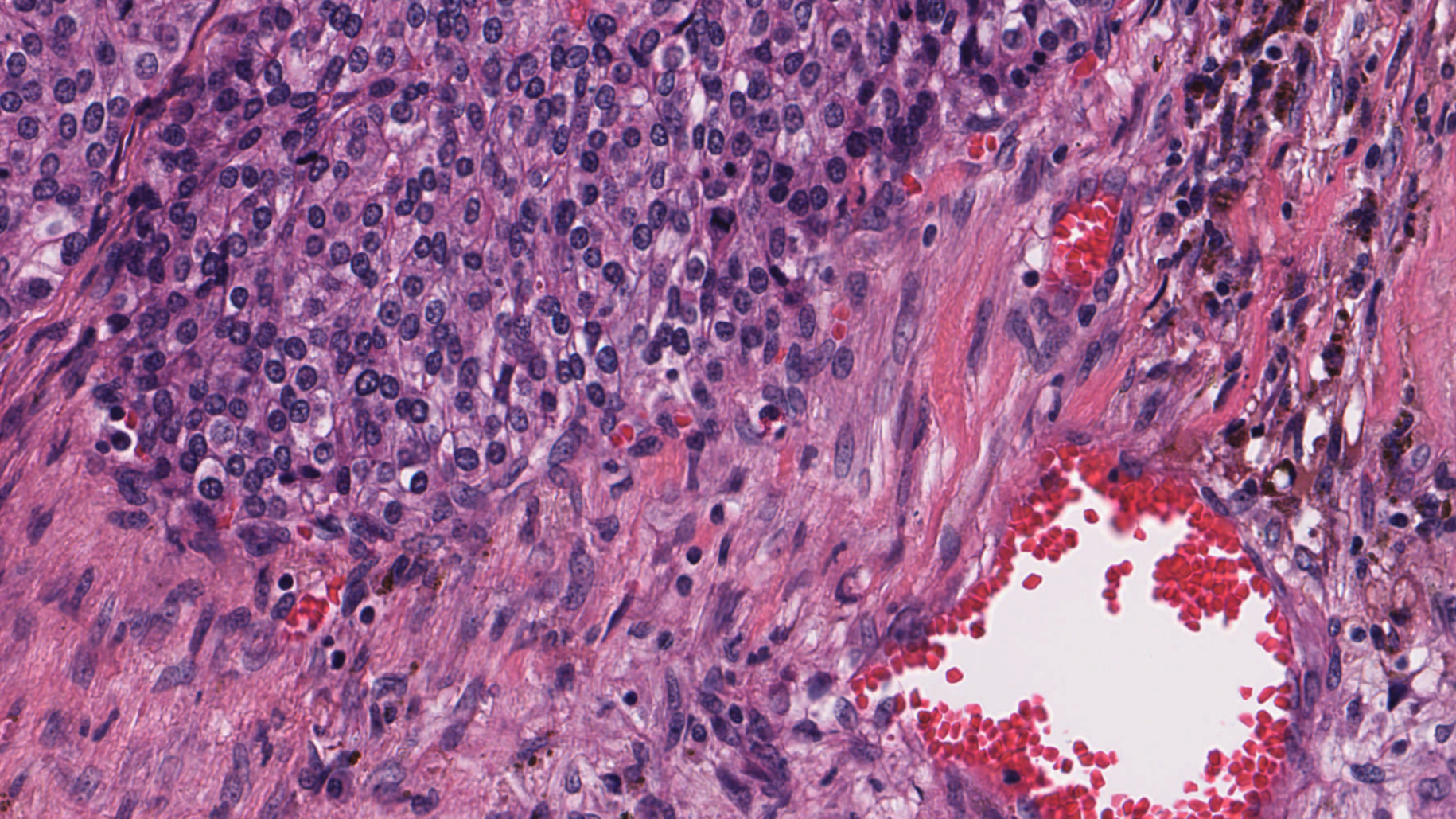
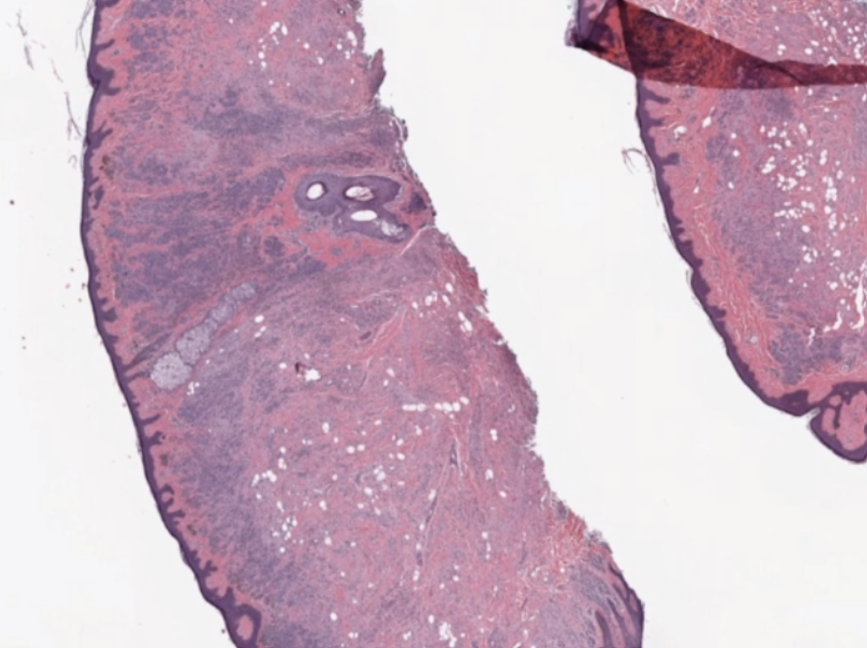
Multidimensional image processing gives you the flexibility to perform functions for traditional two-dimensional filtering in scientific applications. Within medical imaging specifically, computed tomography (CT) and magnetic resonance imaging (MRI) scans require multidimensional image processing to form images of the body and its functions. For example, multidimensional dimensional image processing is used in medical imaging to detect cancer or estimate tumor size (Figure 1).
Multidimensional image processing developer challenges
Outside of identifying, acquiring, and storing the image data itself, working with multidimensional image data comes with its own set of challenges.
First, multidimensional images are larger in size than their 2D counterparts and typically of high resolution, so loading them to memory and accessing them is time-consuming.
Second, processing each additional dimension of image data requires additional time and processing power. Analyzing more dimensions enlarges the scope of consideration.
Third, the computer-vision and image-processing algorithms take longer for analyzing each additional dimension, including the low-level operations and primitives. Multidimensional filters, gradients, and histogram complexity grow with each additional dimension.
Finally, when the data is manipulated, dataset visualization for multidimensional image processing is further complicated by the additional dimensions under consideration and quality to which it must be rendered. In biomedical imaging, the level of detail required can make the difference in identifying cancerous cells and damaged organ tissue.
Multidimensional input/output challenges
If you’re a data scientist or researcher working in multidimensional image processing, you need software that can make data loading and handling for large image files efficient. Popular multidimensional file formats include the following:
- NumPy binary format(.npy)
- Tag Image File Format (TIFF)
- TFRecord (.tfrecord)
- Zarr
- Variants of the formats listed above
Because every pixel counts, you have to process image data accurately with all the available processing power available. Graphics processing units (GPU) hardware gives you the processing power and efficiency needed to handle and balance the workload of analyzing complex, multidimensional image data in real time.
cuCIM addresses multidimensional image processing challenges
Compute Unified Device Architecture Clara IMage (cuCIM) is an open-source, accelerated, computer-vision and image-processing software library that uses the processing power of GPUs to address the needs and pain points of developers working with multidimensional images.
Data scientists and researchers need software that is fast, easy to use, and reliable for an increasing workload. While specifically tuned for biomedical applications, cuCIM can be used for geospatial, material and life sciences, and remote sensing use cases.
cuCIM offers 200+ computer-vision and image-processing functions for color conversion, exposure, feature extraction, measuring, segmentation, restoration, and transforms.
cuCIM is capable and fast image-processing software, requiring minimal changes to your existing pipeline. cuCIM equips you with enhanced digital image-processing capabilities that can be integrated into existing pipelines:
You can integrate using either a C++ or Python application programming interface (API) that matches OpenSlide for I/O and scikit-image for processing in Python.
The cuCIM Python bindings offer many commonly used, computer-vision, image-processing functions that are easily integratable and compilable into the developer workflow.
You don’t have to learn a new interface or programming language to use cuCIM. In most instances, only one line of code is added for transferring images to the GPU. The cuCIM coding structure is nearly identical to that used for the CPU, so there’s little change needed to take advantage of the GPU-enabled capabilities.
Because cuCIM is also enabled for GPUDirect Storage (GDS), you can efficiently transfer and write data directly from storage to the GPU without making an intermediate copy in host (CPU) memory. That saves time on I/O tasks.
With its quick set-up, cuCIM provides the benefit of GPU-accelerated image processing and efficient I/O with minimal developer effort and with no low-level compute unified device architecture (CUDA) programming required.
Free cuCIM downloads and resources
cuCIM can be downloaded for free through Conda or PyPi. For more information, see the cuCIM developer page. You’ll learn about developer challenges, primitives, and use cases and get links to references and resources.