Whole genome sequencing has become an important and foundational part of genomic research, enabling researchers to identify genetic signatures associated with diseases, differentiate sequencing errors from biological signals, and better characterize the genomes of various organisms. With the ongoing COVID-19 pandemic threatening the globe, characterizing, and understanding genomes is now more crucial than ever. Commercially available, next-generation sequencing platforms allow researchers to decode an entire human genome in less than a day. This helps in understanding the susceptibility with infection by SARS-CoV-2; can be used as the basis for vaccine creation; and can be used for therapy selection for an individual based upon their unique genetic signatures, along with many other use cases.
Traditionally, sequencing a whole human genome takes multiple days coupled to a heavy compute power using CPU resources. The Genome Analysis Toolkit (GATK), developed by the Broad Institute, is a multi-purpose software suite for analyzing DNA– and RNA-based sequence data with the primary goal to identify genetic variants. It is generally considered the industry standard toolset. The GATK suite contains several tools, including the Base Quality Score Recalibration (BQSR), Burrows Wheeler Aligner Maximal Effect Match (BWA MEM) for aligning and calibrating genomes, and HaplotypeCaller, which efficiently identifies variants in sequences. These tools are all run as sub-stages in the germline pipeline that identifies germline variants and is one of the algorithms used in this post.
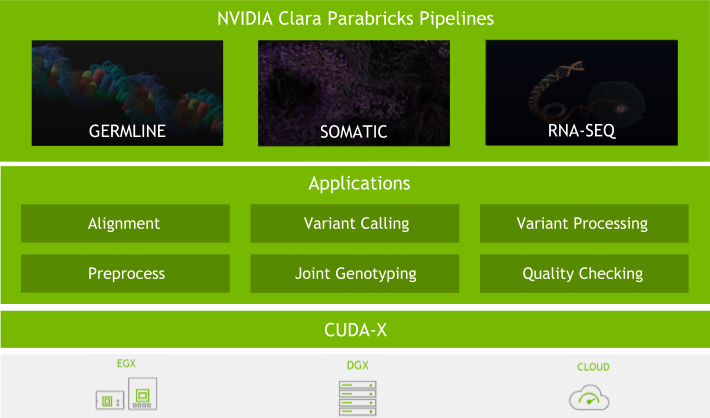
NVIDIA Clara Parabrick Pipelines replicate the functionality of GATK while harnessing the power of GPUs to provide the fastest method to analyze genomes. The software, leveraging the NVIDIA CUDA libraries to accelerate key algorithms, dramatically reduces the time required to analyze a sequence compared to the CPU-only GATK tools and also includes the DeepVariant algorithm developed by researchers at Google. Combined with the power of Amazon Web Services (AWS) and NVIDIA storage partner WekaIO (Weka), the high-performance sequence analysis of NVIDIA Clara Parabricks integrates with the convenience of the cloud to offer a robust, fast, and simple solution to enable faster genomic research over existing state-of-the-art CPU-based solutions. It offers over 33x improvements in performance with over 99.9% concordance of results.
This post is an overview directed at genomic researchers, data scientists, and the IT teams supporting them, the latter of whom are responsible for creating architectures to run sequence analysis jobs. In this post, we walk through the rationale for various components as well as the creation of the Weka filesystem in the cloud using AWS. The solution sets up clients to run the germline variant calling pipeline using Parabricks and various optimizations for both the storage and GPUs to provide the best performance. We also offer an analysis of all results gathered during the tests.
Weka File System
Weka is a modern parallel file system built on NVMe, boasting high throughput and low latencies in an easy-to-manage package. Not only does Weka provide on-premises solutions through OEM storage vendors, they also support cloud deployments and hybrid environments, integrating seamlessly with AWS. Weka was a natural choice for this work as they target many industries, including artificial intelligence, life sciences, and financial services. Given requirements for large shared storage, low latencies, and the ability to work in the cloud, Weka is well-positioned to support those trying to rapidly analyze sequence data. Above all else, Weka is simple to set up and use.
Hybrid model and cloud bursting
Given that this was our first introduction to Parabricks Pipelines, we began experimenting with the pipelines and pulling data on a local server in the lab until we were comfortable with the process. This allowed us to verify the functionality of all tools and validate the dataset before scaling up to larger tests. In the lab, we used an on-premises Weka cluster to store all data and were able to easily archive all data in an Amazon S3 bucket, allowing us to pull the data later on from any cluster with access to AWS servers.
This strategy has several advantages, with one being the ability to experiment on a development machine to get an understanding of how the tools work before running on more expensive or time-sensitive production servers, whether on-premises or in the cloud. Weka makes it easy to transition data to the cloud and tier data to larger, slower storage with a single global namespace. Just a few clicks in Weka’s web interface is all it takes to set up this connection.
CPU baseline
To get a feel for the amount that Parabricks accelerates genome-sequencing performance over GATK, we ran a baseline test on GATK in AWS against an identical workload. The baseline was run against four c5.9xlarge Amazon EC2 instances that use 36 vCPUs with 72 GiB of memory. After running the germline pipeline against the same dataset, it took 33 hours to analyze a single genome, which is over 33x longer than it took Parabricks with four NVIDIA V100 GPUs to perform the equivalent analysis.
Creating the WekaFS file system on AWS
Before starting the germline variant analysis pipeline, you must provision several EC2 instances plus an S3 object store to house both the file system and the GPU clients. You are configuring a Weka file system to store and read the dataset, given that its high-performance characteristics are more than capable of handling this genomics work. Weka has preconfigured scripts at start.weka.io, which perform most of the setup process automatically.
To create a Weka cluster with GPU clients in AWS, go to start.weka.io and fill out the forms based on your requirements. For this specific cluster, use the following settings:
- Weka version 3.7.2
- Total capacity of 10 TiB
- Tiering between SSD and S3
- Instance types of i3en.2xlarge for the storage nodes
- Four p3.8xlarge instances for the clients for 4x NVIDIA V100 GPUs per client
- A custom AMI of “ami-08a5e4d75a3914234” for the clients, which defaults to a supported kernel for Weka version 3.7.2
Configuring Weka
Create either a new filesystem for local storage to be replicated to S3 or rehydrate a filesystem in a new cluster using an existing S3 object store to pre-fill a file system with a copy of the dataset already stored in S3. This makes it easy to deploy new clusters as needed and ensure that no data is lost during the creation and deletion processes.
If the genomic data exists outside of AWS running on Weka, you can create an S3 snapshot from your data to protect and migrate data into AWS. When the data is in AWS, that snapshot can be used to mount the data in a cloud-native version of Weka for use in AWS.
Optimizing performance
Before installing Parabricks and launching tests, make several tweaks to optimize the performance of the pipeline and finish analyzing genomes more rapidly. These optimizations include tuning the GPUs for maximum performance and pre-fetching data from the object store to ensure that data is stored in flash.
Tuning the GPUs
Amazon explains how to optimize GPUs for EC2 instances for maximum performance. For the selected p3.8xlarge instances, run the following steps.
Enable the GPU persistence daemon, which ensures that the GPUs are always initialized, even when no clients are connected to the GPUs.
sudo nvidia-persistenced
Set the maximum frequency for the memory and graphics clocks on the GPUs to improve throughput.
sudo nvidia-smi -ac 877,1530
Using Parabricks
Now you have created a filesystem and optimized the clients for maximum performance, it is time to install Parabricks and run the germline pipeline. While results have been collected for both Parabricks version 2.5.3 and 3.0.0 for comparison’s sake, only the steps to install version 3.0.0 are listed in this post. You must also request a license from the Parabricks developer portal.
Installing Parabricks on GPU-accelerated Amazon EC2 nodes
On the Parabricks developer portal, request a free 1-month trial for NVIDIA Clara Parabricks Pipelines. After it is approved, you receive a link to download a bundle that is used to set up and install Parabricks on the EC2 instances. First, download the bundle onto each node using the provided link.
Next, extract the package and run the installer.
cd /tmp/ tar -xvf parabricks.tar.gz cd ./parabricks sudo ./installer.py
The installer displays the EULA and give several prompts. Answer “yes” for all prompts. When the installer is complete, Parabricks is installed and you can finally run pipelines. Verify by checking the version of pbrun installed:
$ pbrun version pbrun: v3.0.0.2
Running the germline pipeline
The first pipeline to run is the commonly used germline variant analysis pipeline, which analyzes sequence data from a human genome or exome. To run the pipeline, navigate to the Weka mount point on the filesystem and download the genome dataset and reference files. Then, call the germline using the following command:
cd /mnt/weka curl --output NA12878xCO_combined_R1.fastq.gz --silent https://pbgenomicsdata.s3.amazonaws.com/Data/NA12878xCO_combined_R1.fastq.gz curl --output NA12878xCO_combined_R2.fastq.gz --silent https://pbgenomicsdata.s3.amazonaws.com/Data/NA12878xCO_combined_R2.fastq.gz curl --output HG38.tar.gz --silent https://pbgenomicsdata.s3.amazonaws.com/Ref/HG38/HG38.tar.gz tar -xvf HG38.tar.gz pbrun germline --ref Ref/Homo_sapiens_assembly38.fasta --in-fq NA12878xCO_combined_R1.fastq.gz NA12878xCO_combined_R2.fastq.gz --knownSites Ref/Homo_sapiens_assembly38.known_indels.vcf --out-bam output.bam --out-variants output.vcf --out-recal-file report.txt
The germline command takes a little over an hour to complete as it goes through the various phases of the pipeline. After it’s complete, the output displays blocks of text indicating the total time to complete each phase, like the following example:
------------------------------------------------------------------------------ || Program: GPU-BWA mem, Sorting Phase-I || || Version: v3.0.0 || || Start Time: Tue Jun 16 18:08:55 2020 || || End Time: Tue Jun 16 18:59:43 2020 || || Total Time: 50 minutes 48 seconds || ------------------------------------------------------------------------------
This generates multiple Variant Calling Files (VCF), which contain information about the variants identified in the genome compared to the reference files, as well as text files with generic pipeline information.
Running the DeepVariant pipeline
The next pipeline is the DeepVariant algorithm from Google, which is praised for being highly accurate in the research community but which requires significant compute resources. This causes many researchers to avoid running the algorithm on CPUs. Running DeepVariant on Parabricks, however, is significantly quicker for analyzing a genome. To run DeepVariant, run the following command from the file system:
pbrun deepvariant --ref Ref/Homo_sapiens_assembly38.fasta --in-bam output.bam --out-variants output.vcf
Results
To measure the overall effectiveness of the solution, you should analyze several key factors, including the performance gains in using the latest version of Parabricks, time difference between running both germline and DeepVariant on CPUs compared to GPUs, and relative accuracy between CPU and GPU solutions.
Time to completion
These results showcase the average time to complete the main phases of the germline and DeepVariant pipelines:
- The total time for the germline pipeline is the sum of the fq2bam alignment and HaplotypeCaller phases.
- The DeepVariant pipeline equals the sum of the fq2bam and DeepVariant.
The fq2bam results indicate how much of a performance improvement version 3.0.0 of the Parabricks software provides over version 2.5.3. These results help showcase the performance gains that can be achieved by upgrading to the latest version of Parabricks. NVIDIA is constantly making improvements to the performance and stability of the Clara Parabricks suite.
| Test Phase | Parabricks 2.5.3 | Parabricks 3.0.0 |
| FQ2BAM | 81 minutes, 29 seconds | 50 minutes, 51 seconds |
| HaplotypeCaller | 16 minutes | 15 minutes, 36 seconds |
| DeepVariant | 38 minutes, 18 seconds | 38 minutes, 31 seconds |
Performance compared to CPUs
While breaking down the completion time of the various phases of the pipelines running on both CPUs and NVIDIA GPUs, the dramatic performance improvements are readily apparent, as seen in the following table. While Parabricks uses the fq2bam aligner, GATK uses BWA MEM for identical results.
| Test Phase | CPU | GPU |
| FQ2BAM/BWA MEM | 879 minutes, 58 seconds | 50 minutes, 51 seconds |
| HaplotypeCaller | 1081 minutes, 58 seconds | 15 minutes, 36 seconds |
| DeepVariant | 389 minutes, 19 seconds | 38 minutes, 31 seconds |
Accuracy
While the performance numbers from Parabricks are certainly beyond impressive, it’s arguably more impressive that the software can achieve these numbers with equivalent accuracy. The Parabricks implementation has greater than 99.9% identity compared to the CPU-optimized pipeline that we ran against the same dataset.
Conclusion
NVIDIA Clara Parabricks—combined with the power of AWS and Weka—provides numerous advantages over existing CPU-optimized solutions, saving time, money, and resources, all without sacrificing the accuracy of the results. Weka made it simple to expand from a local test environment to the cloud, all while keeping the data intact. With a few additional clicks in Weka’s easy-to-use interface, we were able to rehydrate a new filesystem with data from an S3 bucket, a process that took only a few minutes for a few hundred gigabytes of data. This added an extra layer of security and simplicity to managing the dataset and filesystem in case we lost access to any components. Given the comparable performance between on-premises and cloud-based Weka filesystems, you can opt for the solution that best suits your needs.
If you’re looking to accelerate your genomics pipelines, Clara Parabricks provides the same accuracy and analysis capabilities as the CPU-based GATK toolset, while dramatically reducing the time required to analyze a genome by as much as 33x and simplifying the steps required to initiate pipelines.
For more information, see the following resources:
- Accelerate Genomics Discovery in AWS with NVIDIA GPUs (solution brief)
- HOW TO GUIDE: Analyze Genome Sequence Data on AWS with WekaFS and NVIDIA Clara Parabricks (ebook)
Contributions
Thanks to Charla Bunton-Johnson, and Efraim Grynberg from WekaIO along with Darrin Johnson, Robert Sohigian, Abood Quraini, and Mehrzad Samadi from NVIDIA. Without their collective efforts, this work would not have been possible.