By Jason Phang, PhD student at the NYU Center for Data Science
Breast cancer is the second leading cancer-related cause of death among women in the US. However, screening mammograms require radiologists to arduously pore over extremely high-resolution mammography images, looking for features suggestive of cancerous or suspicious lesions.
This appears like an ideal situation to apply deep convolutional neural networks (CNNs), which have proven to be highly successful at image classification tasks. However, there are many challenges associated with effectively applying CNNs to a real-world medical imaging task.
In our newly published work, we adapt deep CNNs to the breast cancer screening pipeline, showing that trained models can not only perform comparably to radiologists in breast cancer screening exam classification but further improve their performance when combining their expertise with our model’s predictions.
Challenges in Applying Neural Networks to Medical Imaging
The success of deep learning methods in computer vision was built in large part upon large image datasets such as ImageNet, which consists of over 14 million images. However, public breast cancer datasets are fairly small. For example, the Digital Database for Screening Mammography (DDSM), contains only about 10,000 images. To address this, we first constructed the NYU Breast Cancer Screening Dataset, a massive dataset of screening mammograms, consisting of over 1 million mammography images.
The second challenge is that mammography images are of extremely high resolution–much larger than those in ImageNet. Prior work has shown that retaining that high resolution is necessary for achieving the best predictive performance. This places severe constraints on the size of the model architectures we can use based on GPU memory limitations.
To address this, we created a 22-layer ResNet model (which is relatively shallow compared to modern ImageNet models), that more aggressively pooled representations in the earlier layers. Because a full screening mammogram consists of four mammogram images (left/right breasts, and so-called CC/MLO views), we apply four such ResNets for a given exam. Using a shallower architecture means that the model has relatively less modeling capacity, and may not be able to pick up small, localized visual features that could be indicative of cancer.
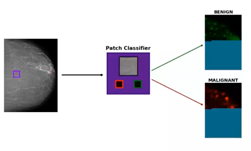
To complement our ResNet model that processes whole mammography images at once, we also trained a full ResNet-50 model on classifying patches, rather than full mammograms. Using lesion segmentations on a subset of our data provided by radiologists, we sample 224×224 patches from our mammogram images and train the patch classification model on determining the presence or absence of benign or malignant lesions in the patch. This produces a very strong but very local cancer classification model. We then apply the patch classification model in a sliding window fashion across our full mammogram and extract the output logits, effectively creating a “heatmap” of local predictions across the mammogram.
By concatenating the patch classification heatmaps with the original mammogram image and training a cancer classification network on the concatenated inputs, our model is able to exploit both global and local features in the mammogram in making a prediction.
The final challenge is that there are relatively few positive examples of cancer in our data set, which is representative of the general screening population. To address this, we first pre-train our model on a proxy task of predicting an initial assessment that radiologists make only based on the screening mammography images—a less decisive label, but one that indicates a larger set of images with suspicious imaging findings. We used the pre-trained weights from the ResNet layers as initialization for our cancer classification model.
We performed our pre-training on four NVIDIA Tesla V100 GPUs for over 100 epochs over the course of two weeks. Training the patch classification took 24 hours on a V100 while generating the patch classification heatmaps for every mammogram image would take approximately 1,000 hours running on a V100, however we are able to parallelize that process across multiple machines. Subsequently, fine-tuning the model for cancer prediction could be effectively performed on a single V100, taking less than a day even for the image-and-heatmaps models.
Deep Neural Networks Improve Radiologists’ Performance in Breast Cancer Screening
We evaluate our cancer classification models in a number of ways, but most pertinently, we compare our models with the performance of radiologists on the task of predicting the presence of cancer-based on screening mammograms. While this task differs somewhat from the work that radiologists normally do, we believe that it is still a useful measure of how our model compares against experts who are deeply familiar with mammograms.
We find that our model tends to outperform an individual radiologist in terms of AUC on the prediction of malignant lesions, but still somewhat underperforms an ensembling of the predictions of multiple radiologists. Moreover, we find that combining our model with the ensembled predictions of radiologists outperforms either! This demonstrates that our model may be identifying aspects of the cancer prediction problem that radiologists may find challenging.
Overall, this demonstrates the capacity for deep neural networks to assist radiologists in their work, potentially alleviating the heavy workload of the number of screening mammograms they read while also improving their accuracy.
Next Steps
Beyond these highly promising results, we are exploring further means of adapting deep neural networks to breast cancer screening, such as object detection methods, utilizing prior screening mammograms, and weakly supervised localization.
This ambitious project is the result of a joint effort between the NYU Center for Data Science and NYU Langone Health. More details about our model and results can be found in our manuscript. Details about the creation of the NYU Breast Screening Dataset can be found in our technical report. We have released code for generating predictions as well as our trained weights on our GitHub.
This article is a guest post written by Jason Phang, a PhD student at the New York University Center for Data Science.