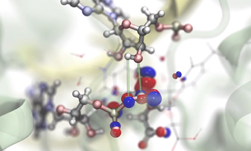
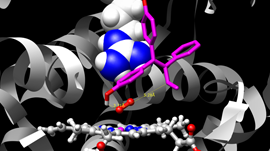
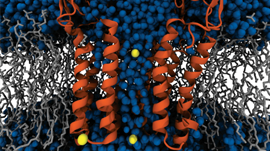
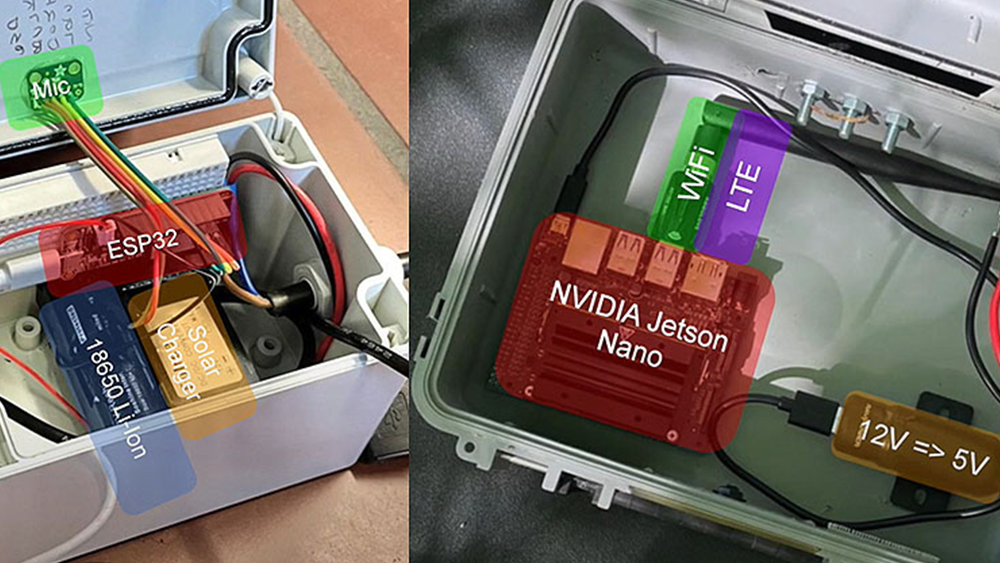
Researchers at the Beckman Institute, part of The University of Illinois, Urbana-Champaign are developing research tools like NAMD and VMD for computational scientists all over the world to analyze large and cumbersome datasets to visualize and simulate molecular dynamics. NAMD is a molecular dynamics simulation package that runs on anything from desktops all the way to supercomputers, and VMD is a molecular visualization, analysis, and simulation preparation tool.
Experimental sciences deliver high-resolution atomic structures for biological complexes, but researchers need to refine those structures, prove their accuracy, and simulate their dynamics while retaining all of the information that makes simulations meaningful and realistic. The amount of data required to achieve the desired results is large and difficult to work with, which is why the Beckman Institute relies on the combination of CUDA and GPUs.
In the video above, John Stone, Senior Research Programmer at the Beckman Institute, explains how CUDA serves as the programming abstraction allowing his team to leverage the GPU compute power.
CUDA does this by allowing researchers to describe hundreds of thousands to millions of independent, data-parallel work units and write software that executes on those work units, all while achieving peak hardware performance.