The Clara Genomics SDK has been upgraded with high performance analysis algorithms for long read sequencing and early access to deep learning-based processing of short read ATAC sequencing.
Features:
- cudaAligner: Myer’s and Hirchberg’s alignment algorithms
- cudaPOA: Python API and optimized implementations of partial order alignment (POA) algorithms including lower memory utilization and additional speed improvements
- AtacWorks: Early access to deep learning based processing of ATAC sequencing data
- CUDA 9 support
GPU Accelerated Racon
The Racon consensus module for genome assembly enabled GPU- accelerated polishing of long reads in v1.4 with the integration of the cudaPOA module in the Clara Genomics SDK. We investigated the performance of Racon with the new cudaAligner and cudaPOA features in this release and were able to achieve significant acceleration.
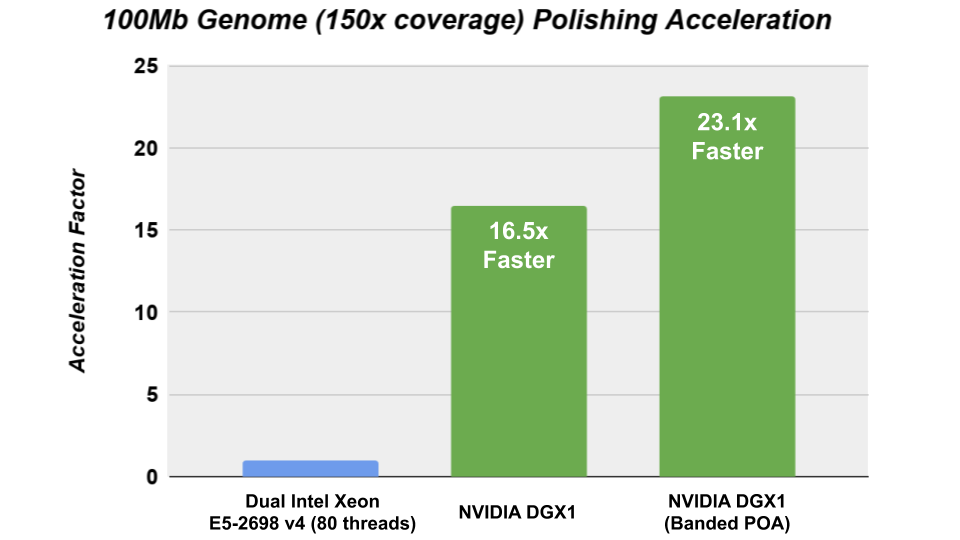
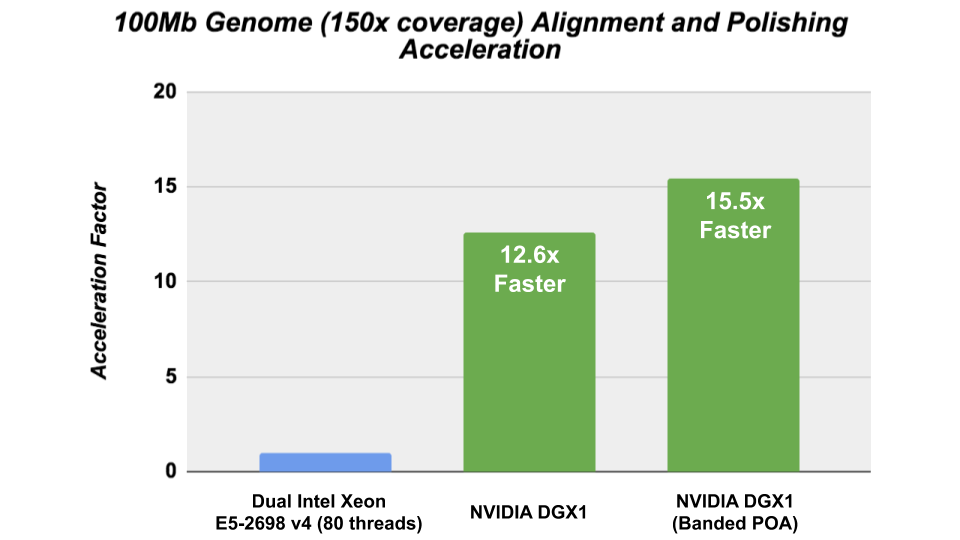
The CPU performance was obtained on Ubuntu 16.04 using the gcc 7.4.0 compiler and the following CXX_FLAGS: – -Wall -Wextra -pedantic -O3 -DNDEBUG -march=broadwell -std=c++11.
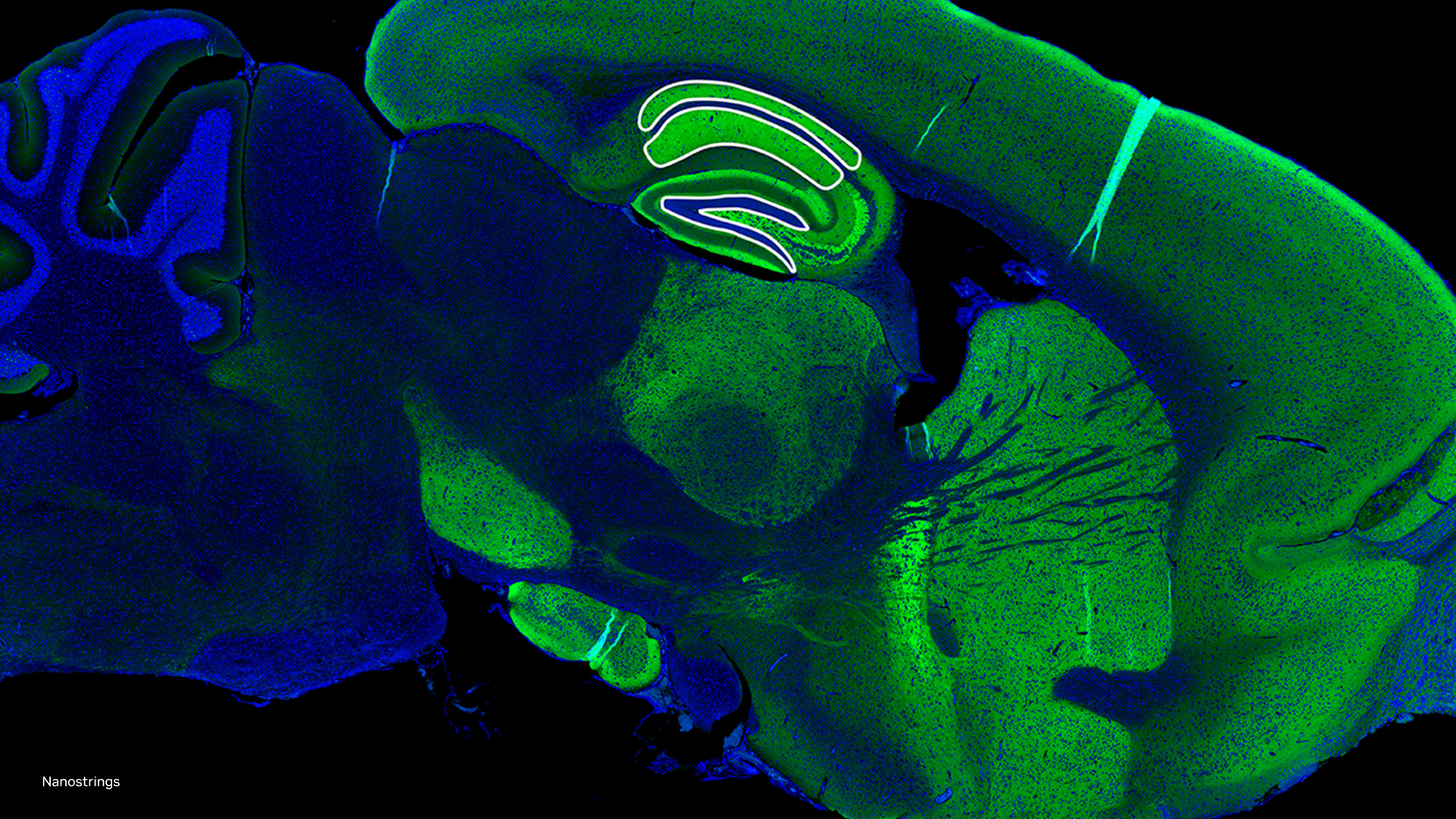
Clara Genomics was created to address the growing size and complexity of genomics sequencing & analysis with accelerated and intelligent computing. The AtacWorks module extends the Clara Genomics SDK to the analysis of short read sequencing data with deep learning; we will share more details about this capability at NVIDIA’s ASHG 2019 workshop on accelerated computing and AI for Genomics.
“We think these tools for ATAC-seq and single-cell ATAC-seq will be useful for individual labs and large-scale studies, providing faster and more accurate analysis of epigenetic changes across cell types” – Jason Buenrostro, Harvard faculty and inventor of ATAC sequencing technologies.