University of Notre Dame researchers developed a deep learning-based system that can accurately determine bond energies.
“Neural networks can be used to make quantitative models of chemical concepts that are not possible with just quantum mechanics,” explains John Parkhill, Assistant Professor of Chemistry & Biochemistry at the University of Notre Dame in Indiana and co-author of the paper. “[Before], I got asked all the time: ‘How much stronger is this bond?’ […] Now I can answer that question, because my machine learnt chemical bonding concepts.”
Using Tesla K80 GPUs and the cuDNN-accelerated TensorFlow deep learning framework, Parkhill and his team trained their neural network on a database of over 130,000 molecules. The trained Bonds-in-Molecules Neural Network (BIM-NN) is able to make predictions of relative bond strengths as well as a trained synthetic chemist.
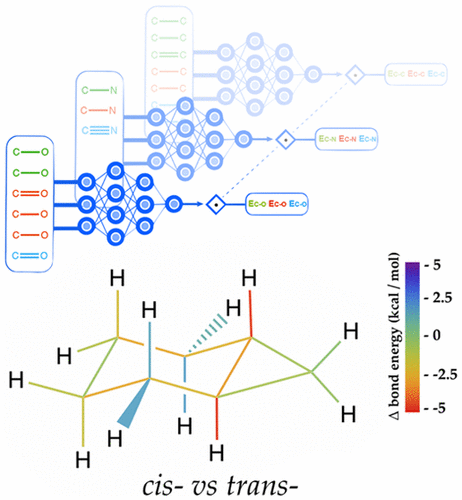
“[Our network] predicts [data] quantitatively and reproducibly. It saves chemists from the impossible tedium of predicting an energy a billion times,” said Parkhill in regards to their software replacing trained chemists.
Read more >
Related resources
- GTC session: The Transformational Role of AI in Computational Chemistry and Molecular Design
- GTC session: Combining Quantum-Based Models With Machine Learning Accelerates Drug Discovery
- GTC session: Combining Machine Learning With Quantum Computing for a New Generation of Quantum Algorithms
- NGC Containers: TorchANI
- Webinar: Isaac Developer Meetup #2 - Build AI-Powered Robots with NVIDIA Isaac Replicator and NVIDIA TAO
- Webinar: Bringing Drugs to Clinics Faster with NVIDIA Computing