“Meet the Researcher” is a series in which we spotlight different researchers in academia who are using GPUs to accelerate their work. This week, we spotlight Jean-Philip Piquemal, Professor at Sorbonne University in Paris, France.
Jean-Philip Piquemal is Professor of Theoretical Chemistry at Sorbonne University and Director of the Theoretical Chemistry Laboratory (LCT), a joint research center with The French National Centre for Scientific Research (CNRS).

He is also an Adjunct Professor of Biomedical Engineering at the University of Texas in Austin. His research is devoted to methodological developments in multiscale quantum chemistry for large systems including new generation polarizable force fields, and Quantum Chemical Topology approaches.
What are your research areas of focus?
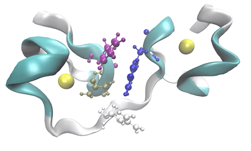
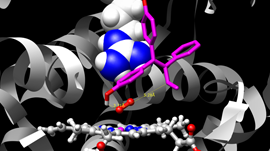
My team develops the massively parallel Tinker-HP simulation package dedicated to new generation polarizable force fields molecular dynamics. Our research is focused on methodological developments for multiscale quantum chemistry methods and new generation (polarizable) force fields able to capture many-body physics. We extensively use High Performance Computing to tackle large scale applications of these techniques to biological systems including metalloproteins and viruses, chemical reactivity and drug design.
What motivated you to pursue this research area?
I have always been interested in interdisciplinary research. Indeed, working at the interface of theoretical chemistry, biophysics, HPC and applied mathematics is quite exciting as it allows to push the limits of what it is possible to simulate in term of complex systems.
Tell us about your current research projects.
Our long standing project is the development of the massively parallel Tinker-HP software which is aimed at stimulating very large complex system ranging from biomolecules to ionic liquids. It is especially designed to be able to tackle efficiently new generation polarizable force fields molecular dynamics (MD) simulations. We are also interested in hybrid QM/MM MD simulations. We are using HPC to make these approaches scalable in order to apply them to a variety of applications and challenges.
What problems or challenges does your research address?
Our research tries to address the challenges of accuracy and efficiency in molecular simulations. How to transfer as much as possible of the accuracy of quantum physics into simulations. Thanks to high performance computing, the capability to sample large systems using molecular dynamics.
What is the (expected) impact of your work on the field/community/world?
We expect that our work will improve the predictability of molecular dynamics simulations especially for complex biomolecules (including metalloproteins), charged systems such ionic liquids or heavy metal complexes. We hope to be able to improve significantly free energy simulations for drug discovery.
How have you used NVIDIA technology either in your current or previous research?
The NVIDIA GPUs acceleration technology coupled to our Tinker-HP HPC setup allow us to cumulate the use of several GPUs to power up our high resolution simulations. Such enhanced capabilities make possible to tackle systems which were out of reach of the past implementations.
Did you achieve any breakthroughs in that work?
Our multi-GPUs powered Tinker-HP is able to perform longer simulations on larger systems using high resolution polarizable force fields. It will certainly solve many problem but for now, its phase advance version has been mobilized against the COVID-19 virus where its extended capabilities are used to model the various viral proteins and to perform free energy simulation of potential drugs.
What’s next for your research?
The exascale era is opening many challenges but first, we’ll definitely try to push further the limits of our methodology on these new supercomputers towards simulating larger and more realistic biosystems such as cells and viruses models.
Any advice for new researchers?
Get interest in other disciplines, especially HPC and applied mathematics. It is certainly not the easiest path but interdisciplinary is the key.
Join Jean-Philip Piquemal on Tuesday, June 23rd in NVIDIA’s Compute4Covid webinar. The webinar is called Using GPU-Powered Tinker-HP at Sorbonne University for COVID-19 Research. During the webinar, you can learn how the Tinker-HP simulation package is advancing COVID-19 research. Register now.