The C++ standard library contains a rich collection of containers, iterators, and algorithms that can be composed to produce elegant solutions to complex problems. Most importantly, they are fast, making C++ an attractive choice for writing highly performant code.
NVIDIA recently introduced stdpar: a way to automatically accelerate the execution of C++ standard library algorithms on GPUs using the nvc++ compiler. This means that C++ programs using the standard library containers and algorithms can now run even faster.
In this post, I explore a way to bring GPU-accelerated C++ algorithms to the Python ecosystem. I use Cython as a way to call C++ from Python and show you how to build Cython code with nvc++. I present two examples: a simple task of sorting a sequence of numbers and a more complex real-world application, the Jacobi method. In both cases, you’ll see impressive performance gains over the traditional approach of using NumPy. Finally, I discuss some current limitations and next steps.
Using C++ from Python
If you’ve never used Cython before or could use a refresher, here’s an example of writing a function in Cython that sorts a collection of numbers using C++’s sort function. Place the following code in a file, cppsort.pyx:
# distutils: language=c++ from libcpp.algorithm cimport sort def cppsort(int[:] x): sort(&x[0], &x[-1] + 1)
Here is an explanation of the different parts of this code:
- The first line tells Cython to use C++ mode as opposed to C (the default).
- The second line makes the C++ sort function available. The keyword
cimportis used to import C/C++ functionality. - Finally, the program defines a function,
cppsort, that does the following:- Accepts a single parameter
x. The precedingint[:]specifies the type ofx, in this case, a one-dimensional array of integers. - Calls
std::sortto sort the elements ofx. The arguments&x[0]and&x[-1] + 1refer to the first and one past the last element of the array, respectively.
- Accepts a single parameter
For more information about Cython syntax and features, see Cython Tutorial.
Unlike .py files, .pyx files cannot be imported directly. To call your function from Python, you must build cppsort.pyx into an extension that you can import from Python. Run the following command:
cythonize -i cppsort.pyx
There are a few things that happen with this command (Figure 1). First, Cython translates the code in cppsort.pyx to C++ and generates the file cppsort.cpp. Next, the C++ compiler (in this case, g++) compiles that C++ code into a Python extension module. The name of the extension module is something like cppsort.cpython-38-x86_64-linux-gnu.so.

The extension module can be imported from Python, just like any other Python module:
In [1]: from cppsort import cppsort
As a test of the function, sort a NumPy array of integers:
In [2]: import numpy as np In [3]: x = np.array([4, 3, 2, 1], dtype="int32") In [4]: print(x) [4 3 2 1] In [5]: cppsort(x) In [6]: print(x) [1 2 3 4]
Fantastic! You’ve effectively sorted an array in Python using C++ std::sort!
GPU acceleration using nvc++
C++ standard library algorithms such as std::sort can be called with an additional parallel execution policy argument. This argument tells the compiler that you want the execution of the algorithm to be parallelized. A compiler such as g++ may choose to parallelize the execution using CPU threads. However, if you compile your code using the nvc++ compiler, and pass the -stdpar option, the execution is accelerated by the GPU. For more information, see Accelerating Standard C++ with GPUs Using stdpar.
Another important change to make is to create a local copy of the input array within the cppsort function. This is because the GPU can only access data that is allocated in code compiled by nvc++ and the -stdpar option. In this example, the input array is allocated by NumPy, which may not be compiled using nvc++.
The following code example is the cppsort function re-written to include the earlier changes. It includes the use of a vector for managing the local copy of the input array, and the copy_n function for copying data to and from it.
from libcpp.algorithm cimport sort, copy_n from libcpp.vector cimport vector from libcpp.execution cimport par def cppsort(int[:] x): cdef vector[int] temp temp.resize(len(x)) copy_n(&x[0], len(x), temp.begin()) sort(par, temp.begin(), temp.end()) copy_n(temp.begin(), len(x), &x[0])
Building the extension with nvc++
When you run the cythonize command, the regular host C++ compiler (g++) is used to build the extension (Figure 1). For the execution of algorithms invoked with the par execution policy to be offloaded to the GPU, you must build the extension using the nvc++ compiler (Figure 2). You also must pass a few custom options, such as -stdpar, to the compiler and linker commands. When the build process involves additional steps such as these, it’s often a good idea to replace the use of the cythonize command with a setup.py script. For more information about a detailed implementation, see the setup.py file in shwina/stdpar-cython GitHub repo, which modifies the build process to use nvc++ along with the appropriate compiler and linker flags.

Using the setup.py file, the following command builds the extension module:
CC=nvc++ python setup.py build_ext --inplace
The way you import and use the function from Python is unchanged, but the sorting now happens on the GPU:
In [3]: x = np.array([4, 3, 2, 1], dtype="int32") In [4]: cppsort(x) # uses the GPU!
Performance
Figure 3 shows a comparison of the cppsort function with the NumPy .sort method. The code used to generate these results can be found sort.ipynb Jupyter notebook. Three versions of the function are presented:
- The sequential version calls
std::sortwithout any execution policy. - The CPU parallel version uses the parallel execution policy and is compiled with g++.
- The GPU version also uses the parallel execution policy, but is compiled with
nvc++and the-stdparcompiler option.
For larger problem sizes, the GPU version performs significantly faster than the others—about 20x faster than NumPy .sort!
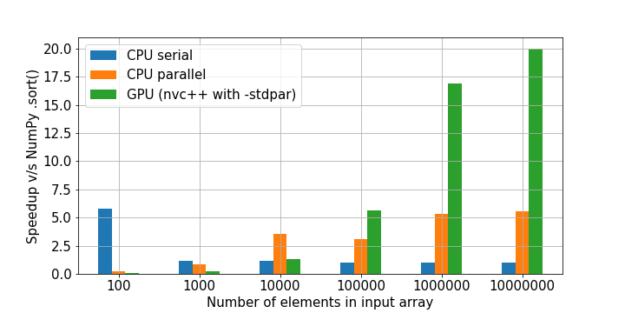
CPU benchmarks were run on a system with an Intel Xeon Gold 6128 CPU. GPU benchmarks were run on an NVIDIA A100 GPU.
Example application: Jacobi solver
As a more complex example, look at using the Jacobi method to solve the two-dimensional heat equation. This mathematical equation can be used, for example, to predict the steady-state temperature in a square plate that is heated on one side.
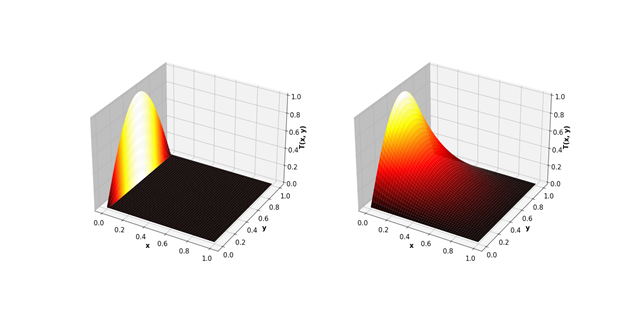
The Jacobi method consists of approximating the square plate with a two-dimensional grid of points. A two-dimensional array is used to represent the temperature at each of these points. Each iteration updates the elements of the array from the values computed at the previous step, using the following update scheme:
This is repeated until convergence is reached: when the values obtained at the end of two subsequent iterations do not differ significantly.
From a programming standpoint, the Jacobi method can be implemented in C++ using the standard library algorithms std::for_each for performing the update step, and std::any_of for checking for convergence. The following Cython code uses these C++ functions to implement a Jacobi solver. For more information about the implementation, see the jacobi.ipynb Jupyter notebook.
# distutils: language=c++ # cython: cdivision=True from libcpp.algorithm cimport swap from libcpp.vector cimport vector from libcpp cimport bool, float from libc.math cimport fabs from algorithm cimport for_each, any_of, copy from execution cimport par, seq cdef cppclass avg: float *T1 float *T2 int M, N avg(float* T1, float *T2, int M, int N): this.T1, this.T2, this.M, this.N = T1, T2, M, N inline void call "operator()"(int i): if (i % this.N != 0 and i % this.N != this.N-1): this.T2[i] = ( this.T1[i-this.N] + this.T1[i+this.N] + this.T1[i-1] + this.T1[i+1]) / 4.0 cdef cppclass converged: float *T1 float *T2 float max_diff converged(float* T1, float *T2, float max_diff): this.T1, this.T2, this.max_diff = T1, T2, max_diff inline bool call "operator()"(int i): return fabs(this.T2[i] - this.T1[i]) > this.max_diff def jacobi_solver(float[:, :] data, float max_diff, int max_iter=10_000): M, N = data.shape[0], data.shape[1] cdef vector[float] local cdef vector[float] temp local.resize(M*N) temp.resize(M*N) cdef vector[int] indices = range(N+1, (M-1)*N-1) copy(seq, &data[0, 0], &data[-1, -1], local.begin()) copy(par, local.begin(), local.end(), temp.begin()) cdef int iterations = 0 cdef float* T1 = local.data() cdef float* T2 = temp.data() keep_going = True while keep_going and iterations < max_iter: iterations += 1 for_each(par, indices.begin(), indices.end(), avg(T1, T2, M, N)) keep_going = any_of(par, indices.begin(), indices.end(), converged(T1, T2, max_diff)) swap(T1, T2) if (T2 == local.data()): copy(seq, local.begin(), local.end(), &data[0, 0]) else: copy(seq, temp.begin(), temp.end(), &data[0, 0]) return iterations
Performance
Figure 5 shows the speedups obtained from three different C++-based Cython implementations compared to a NumPy implementation of Jacobi iteration. The code for generating these results can also be found in the earlier notebook.
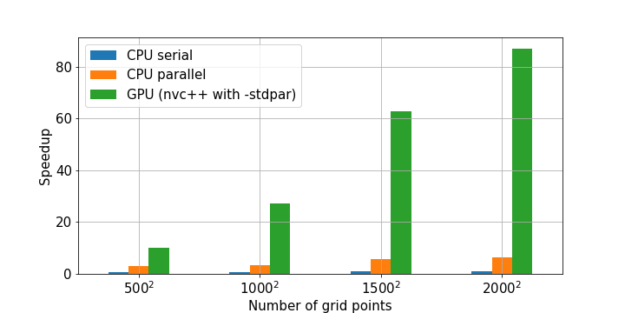
Cython and stdpar bring accelerated algorithms to Python
stdpar introduced a way for C++ standard library algorithms such as counting, aggregating, transforming, and searching to be executed on the GPU. With Cython, you can use these GPU-accelerated algorithms from Python without any C++ programming at all.
Cython interacts naturally with other Python packages for scientific computing and data analysis, with native support for NumPy arrays and the Python buffer protocol. This enables you to offload compute-intensive parts of existing Python code to the GPU using Cython and nvc++. The same code can be built to run on either CPUs or GPUs, making development and testing easier on a system without a GPU.
One important current limitation of this approach is that a copy of the input data is often required. In Python, memory is allocated inside libraries such as NumPy. Such externally allocated memory cannot be accessed by the GPU. It’s also important to keep in mind the limitations mentioned in a previous post, Accelerating Standard C++ with GPUs Using stdpar.
Where to go next?
Here’s how to get started using Cython and nvc++ together:
- Install the NVIDIA HPC SDK. You need a minimum version of 20.9.
- Follow the instructions in the README and run the example notebooks in this shwina/stdpar-cython GitHub repo.
Get in touch!
This post represents some early explorations of using Cython and nvc++ together. I’m excited for you to try it yourself and to hear about the problems that you solve! Start a discussion or report any problems you might encounter by posting to the NVIDIA Developer Forum.