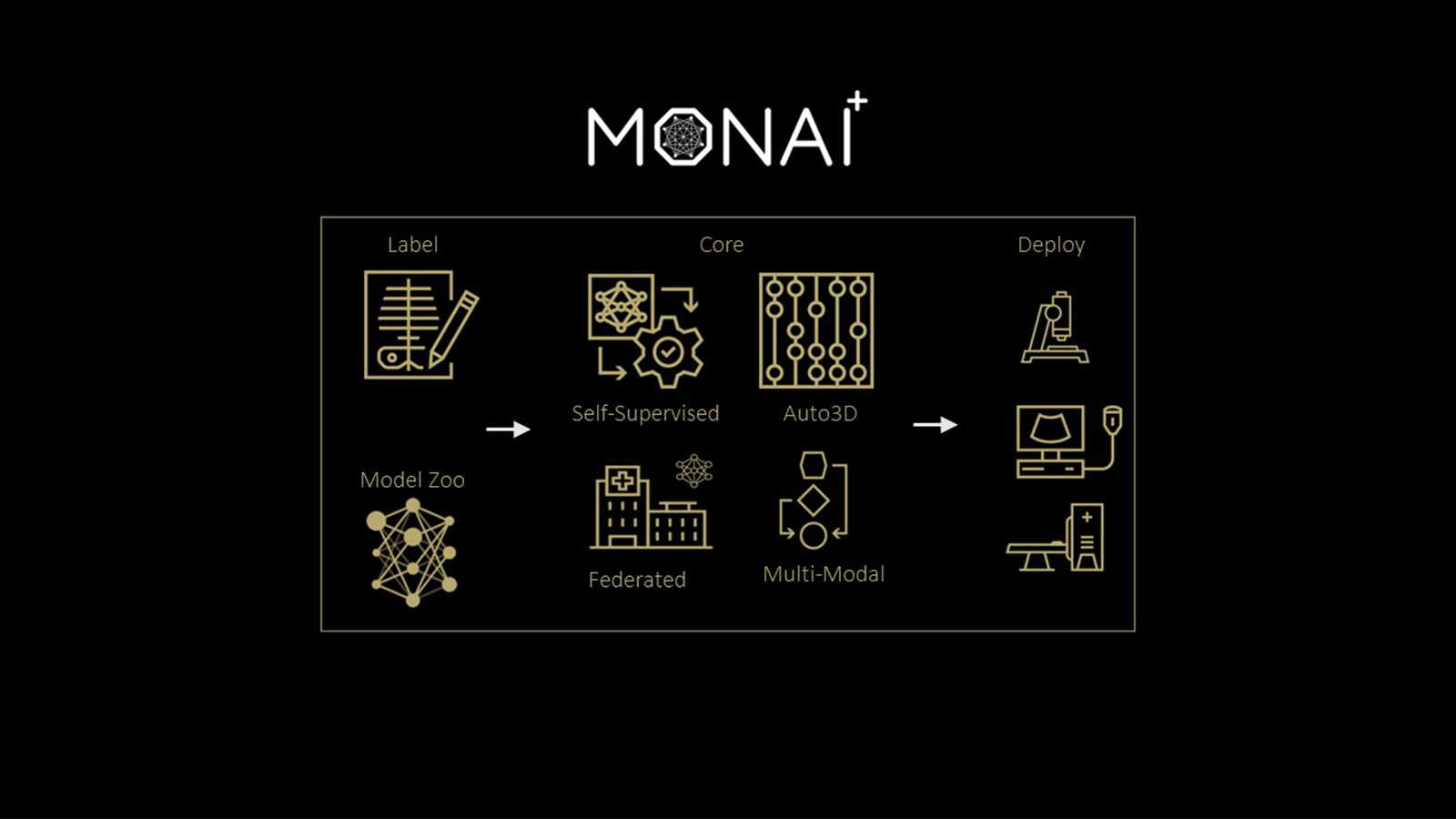
Deep learning models have been successfully used in medical image analysis problems but they require a large, curated amount of labeled images to obtain good performance. Creating such annotations are tedious, time-consuming and typically require clinical expertise.
To address this gap, Project MONAI has released MONAI Label v0.1 – an intelligent open source image labeling and learning tool that helps researchers and clinicians collaborate, create annotated datasets easily and quickly, and build AI models in a standardized MONAI paradigm.
MONAI Label enables the adaptation of AI models to the clinical task at hand by continuously learning from the user’s interactions and new labels. It powers an AI Assisted annotation experience, allowing researchers and developers to make continuous improvements to their applications with iterative feedback from clinicians who are typically the end users of the medical imaging AI models.
At the Children’s Hospital of Philadelphia (CHOP), Dr. Matthew Jolley explains how they are innovating and driving clinical impact with machine learning algorithms.
“Children with congenital heart disease demonstrate a broad range of anatomy, and there are few readily available tools to facilitate image-based structural phenotyping and patient specific planning of complex cardiac interventions. However, currently 3D image-based heart model creation is slow, even in the hands of experienced modelers. As such, we have been working to develop machine learning algorithms to create models of heart valves in children with congenital heart disease, such as the tricuspid valve in hypoplastic left heart syndrome. Ongoing development of automation based on machine learning will allow rapid modeling and precise quantification of how a dysfunctional valve differs from normal valves across multiple parameters. That “structural valve profile” for an individual can then be contextualized within the spectrum of anatomy and function we see in the population, which may eventually inform improved medical decision making and interventions for children.”
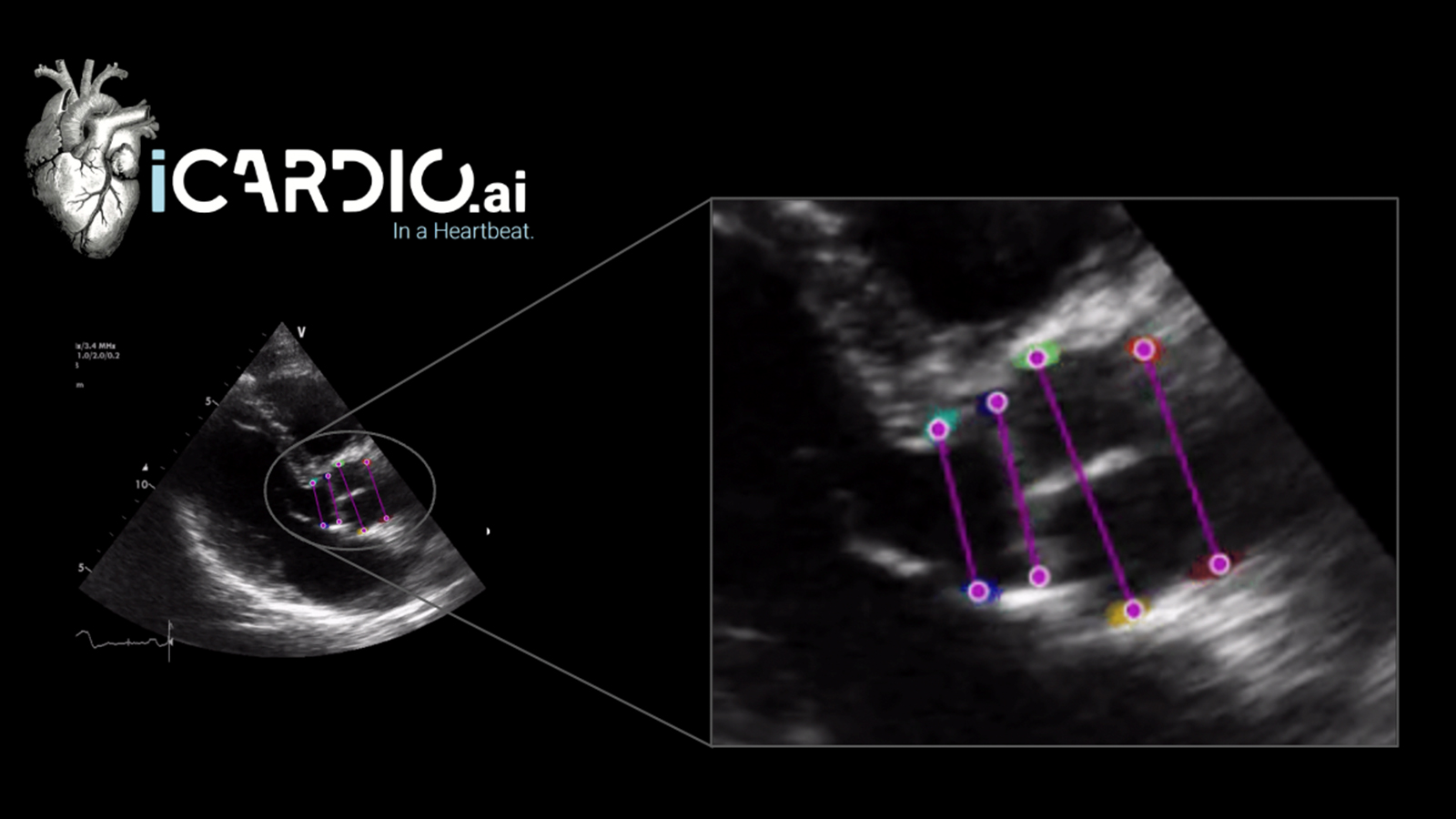
With MONAI Label we envision creating a community of researchers and clinicians like Dr. Jolley and his team who can build upon a well maintained software foundation that will accelerate collaboration through continuous learning. The MONAI Label team and CHOP collaborated through a Slicer week project, and successfully developed a MONAI Label application for leaflet segmentation of heart valves in 3D echocardiographic (3DE) images. The team is now working to deploy this model as a MONAI Label application on a public facing server at CHOP where clinicians can directly interface with the model and trigger a training loop for adaptation – learn more.
It is incredibly important for an open source initiative like Project MONAI to have clinicians in the loop as we converge to develop a common set of best practices for AI lifecycle management in healthcare imaging. To quote Dr. Jolley:
“Open-source frameworks like Project MONAI provide a standardized, transparent, and reproducible template for the creation of, and deployment of medical imaged-focused machine learning models, potentiating efforts such as ours. They allow us to focus on investigating novel algorithms and their application, rather than developing and maintaining software infrastructure. This in turn has accelerated research progress which we are actively translating into tools of practical relevance to the pediatric community we serve.”
WHAT IS INCLUDED IN MONAI LABEL V0.1
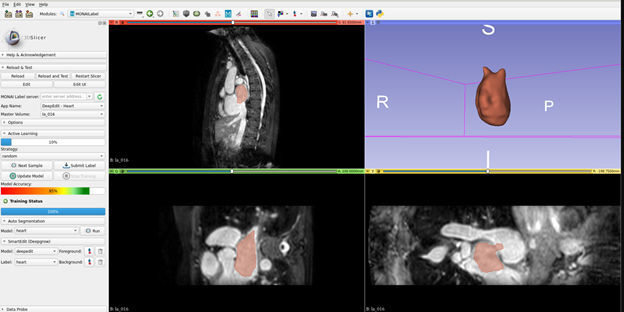
MONAI Label is an open-source server-client system that is easy to set up and can run locally on a machine with one or two GPUs. The initial release does not yet support multiple user sessions, therefore both server and client operate on the same machine.
MONAI Label delivers on MONAI’s core promise of being modular, Pythonic, extensible, easy to debug, user friendly, and portable.
MONAI v0.1 includes:
- MONAI Label server: REST API server that facilitates communication with the viewer clients i.e. (Slicer, OHIF, etc).
- MONAI Label sample applications that can be adapted to a given clinical task for MR/CT modality including DeepGrow and DeepEdit which are developed by MONAI researchers.
- With v0.1, we have released a 3DSlicer plugin for MONAI Label to kick start users in the MONAI Label experience
Future releases of NVIDIA Clara AIAA will also leverage the MONAI Label framework. We continue to bring together development efforts for NVIDIA Clara medical imaging tools and MONAI to deliver domain-optimized, robust software tools for researchers and developers in healthcare imaging.
With contributions from an engaged community, MONAI Label aims to reduce the cost of labeling and maximize the collaboration between researchers & clinicians. Get started today with sample applications available on the MONAI Label GitHub and follow along with our step-by-step getting started guide available in the MONAI Label Documentation.