Case Study
Scaling Multimodal Scientific Literature Understanding with NVIDIA Nemotron Parse
.jpg)
About Edison Scientific: Edison Scientific built Kosmos, an autonomous AI scientist that accelerates scientific discovery by compressing six months of expert analysis into a single day of compute. The company works with universities, national labs, and healthcare organizations, serving more than 50,000 users worldwide.
Challenge
What Edison Scientific needed to do
Edison Scientific needed to enable 50,000 scientists to autonomously synthesize large volumes of scientific literature, perform data analysis, and integrate findings into new hypotheses. To support this objective, the team developed Kosmos, an agentic AI scientist implemented as a multi-agent system capable of executing long-running workflows over 24-hour periods.
Within Kosmos, the Literature sub-agent is responsible for retrieving and understanding scientific papers at scale. Its role is to extract and cite key evidence, including figures, tables, and experimental results, that informs downstream reasoning and hypothesis generation.
Constraints
Cost efficiency and retrieval accuracy: At scale, extraction failures directly impact system reliability and user trust. Improperly cropped figures or missing axes can invalidate otherwise correct answers. The target experience requires responses grounded in correctly extracted and cited figures or tables, paired with accurately cropped visual evidence, while maintaining cost efficiency across thousands of papers per request.
Multimodal document diversity: Scientific papers embed critical information in non-standardized visual elements such as multi-panel figures, irregular tables, equations, and process diagrams. These elements are often not restated in surrounding text, making accurate visual extraction a requirement rather than an optimization.
Infrastructure: Rule-based PDF parsers struggle to preserve spatial relationships between document elements, particularly for panel figures, flow diagrams, and tables with irregular structure or embedded images. When spatial context is lost, semantic interpretation becomes unreliable, leading to incomplete or incorrect representations of scientific findings.
Throughput and latency: Achieving high-fidelity literature understanding across thousands of papers requires model-based document parsing capable of high-precision visual and semantic understanding under real-world variability, without introducing unacceptable latency for long-running agentic workflows.
System architecture
To meet the document understanding requirements, Edison integrated a PaperQA reader powered by Nemotron Parse into Literature, a sub-agent of Kosmos. This reader enables Literature to autonomously map scientific papers to researcher-prompted tasks, supporting large-scale retrieval and reasoning workflows.
Nemotron Parse processes documents on a per-page basis, producing classified bounding boxes and parsed text for each page. Parsed text is indexed to support both embedding-based and keyword-based retrieval. Bounding boxes are used to segment figures, tables, and other visual regions, preserving spatial context and enabling downstream multimodal LLM reasoning over visual content.
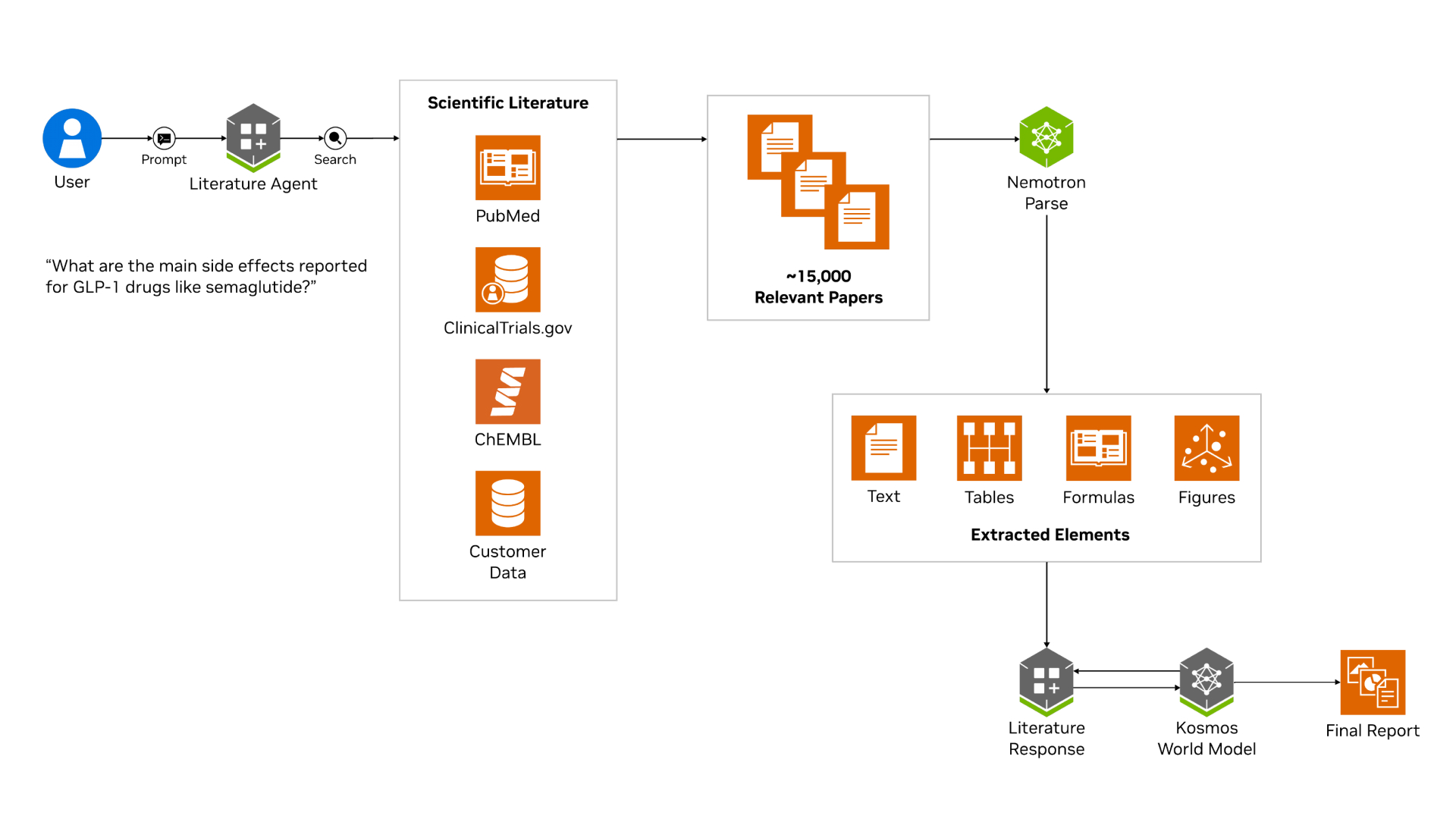
Document processing pipeline overview and flow
Documents are pre-processed once and cached:
Identify and load documents: Documents are first identified and loaded using external metadata providers, including Crossref, Semantic Scholar, and OpenAlex. These sources are used to infer document metadata such as DOI, title, and publication date.
Rasterization: Each document is rasterized by rendering each page as a PNG at 150 DPI. Pages are concurrently screenshotted and sent to Nemotron Parse, ensuring parsing latency does not scale linearly with page count.
Nemotron Parse inference: For each page, Nemotron Parse provides the below.
> Markdown-formatted text content, with tables and formulas represented in LaTeX
> A bounding box corresponding to the region’s spatial location on the page, converted into a PNG image
> One of 13 structural classifications: Text, Title, Section-header, Table, Picture, Formula, Caption, Footnote,
Bibliography, List-item, Page-header, Page-footer, or Table of ContentsDeduplication: To reduce redundant storage, media elements that appear identical across multiple pages, such as journal logos, are deduplicated and stored once rather than replicated on each page.
Chunking: Parsed text is then split into chunks, with associated media linked to the relevant text segments. When media spans chunk boundaries, it is associated with each adjacent chunk to preserve context.
Media enrichment: Each equation, figure, or table is passed to a VLM to generate a detailed scientific description, with surrounding text as context. This enrichment step also identifies non-scientific media, such as logos or decorative elements, which are discarded to prevent context degradation and reduce memory usage.
Embedding: Text embeddings are generated for each text chunk with the (optional) enriched media descriptions.
An area of future exploration is the use of multimodal embeddings to directly encode text and visual content, potentially eliminating the need for a separate media enrichment step.
From there, we enter the core of PaperQA
Context retrieval: Based on the user's query, search for relevant papers. Fetch all cached text chunks and media for the relevant papers.
Context summarization: For each agent-proposed intermediary question, embedding-based retrieval is performed to identify the top K relevant chunks and media. A VLM generates a summary for each retrieved context and assigns a relevance score on a 1-10 scale. Prompts can be expanded to include explicit quotations and references to visual content when applicable.
Answer generation: All ranked contexts are then assembled into a final prompt. The system produces a synthesized response that cites supporting contexts, with optional post-processing formatting citations in MLA style.
Background
The Literature agent builds on PaperQA to retrieve and synthesize scientific literature and is evaluated on LABBench2, a rigorous evaluation setting with open-ended questions that require retrieving the source PDF.
Although modern language models can tolerate suboptimal text extraction, accurate figure extraction is a limiting factor for multimodal tasks such as LABBench2’s figure understanding task FigQA2. Errors in figure parsing, such as missing axes, truncated panels, or loss of spatial context, can prevent the correct interpretation of scientific results.
All results in this case study are reported on LABBench2, reflecting representative usage of Nemotron Parse, part of the NVIDIA Nemotron™ family, for multimodal scientific literature understanding.
Integration details
Key design decisions
Nemotron Parse v1.1 was introduced as the primary PDF parsing component for Literature, replacing previously evaluated rule-based and hybrid parsers.
Media regions segmented by Nemotron Parse are captioned using a VLM. This design allows text embeddings to be generated uniformly for text-only chunks as well as chunks associated with figures, tables, or equations, enabling consistent retrieval across modalities.
The inference stack was deployed on NVIDIA H200 GPUs using Modal Labs to support low-latency, high-throughput parsing and downstream processing.
Deduplicating media and also discarding non-scientific media to keep memory low while optimizing media placed into context.
Nemotron Parse was integrated into a lightweight Python package, paper-qa-nemotron, which connects it to Literature as a PaperQA reader. Evaluations conducted using this reader, described in subsequent sections, demonstrate that Nemotron Parse achieved substantially higher question-answer accuracy than all other parsers evaluated in this system.
What didn’t work initially
Rule-based PDF parsers were unable to robustly handle the variability present in academic literature. These approaches frequently failed to capture critical details in figures while simultaneously inflating context with oversized or poorly cropped visual regions.
In-memory sequential PDF parsing results in parse time that scales linearly with page count. For example, ingesting a 300-page PhD thesis would either slow down the system or be dropped if a preprocessing timeout is required.
Believing one’s system works when testing only quality metrics. There is significant variation across PDFs, which is where Nemotron Parse shines.
Results
Benchmark
LABBench2 is a benchmark released in February 2026 for evaluating the capabilities of AI systems on practical biology research tasks. In this case study, evaluation is conducted using three question-answering tasks drawn from a prerelease version of LABBench2, each targeting a different modality involved in scientific literature understanding:
FigQA2: figure understanding
TableQA2: table understanding
LitQA3: text understanding
LABBench2’s realism and challenge make it a stronger benchmark, so this post uses an early-access version provided by Edison Scientific.
Sample agent trajectory
Here is a sample FigQA2 question targeting DOI 10.1523/JNEUROSCI.2526-13.2014:
A study investigated the physiological consequences of the S218L mutation in CaV2.1 calcium channels on synaptic transmission to identify mechanisms underlying the neurobiological phenotypes of hemiplegic migraine type I. In this work, at what membrane potential is the calcium current amplitude comparable between WT and KI conditions? Round to the nearest multiple of 10.
The ideal answer is -30 mV from Figure 1’s subfigure A2 (view figure below).
To start, Literature runs three paper searches from the years 1990 to 2020 with the following queries:
S218L knock-in CaV2.1 synaptic transmission physiological consequences
familial hemiplegic migraine S218L knockin mouse CaV2.1 calcium current amplitude voltage
Cav2.1 S218L KI calcium currents membrane potential comparable WT
Collectively these searches fetch the target paper alongside 25 others. Assuming about 30 pages per PDF, this results in about 750 pages for Nemotron Parse. For each page:
Capture each page as a 596px by 842px RGB screenshot, roughly equal to 1470KB in size.
Passing the screenshots to Nemotron Parse’s markdown_bbox tool, which segments bounding boxes and provides the text contents of each box, if relevant. An example segmentation can be shown in Figure 1.
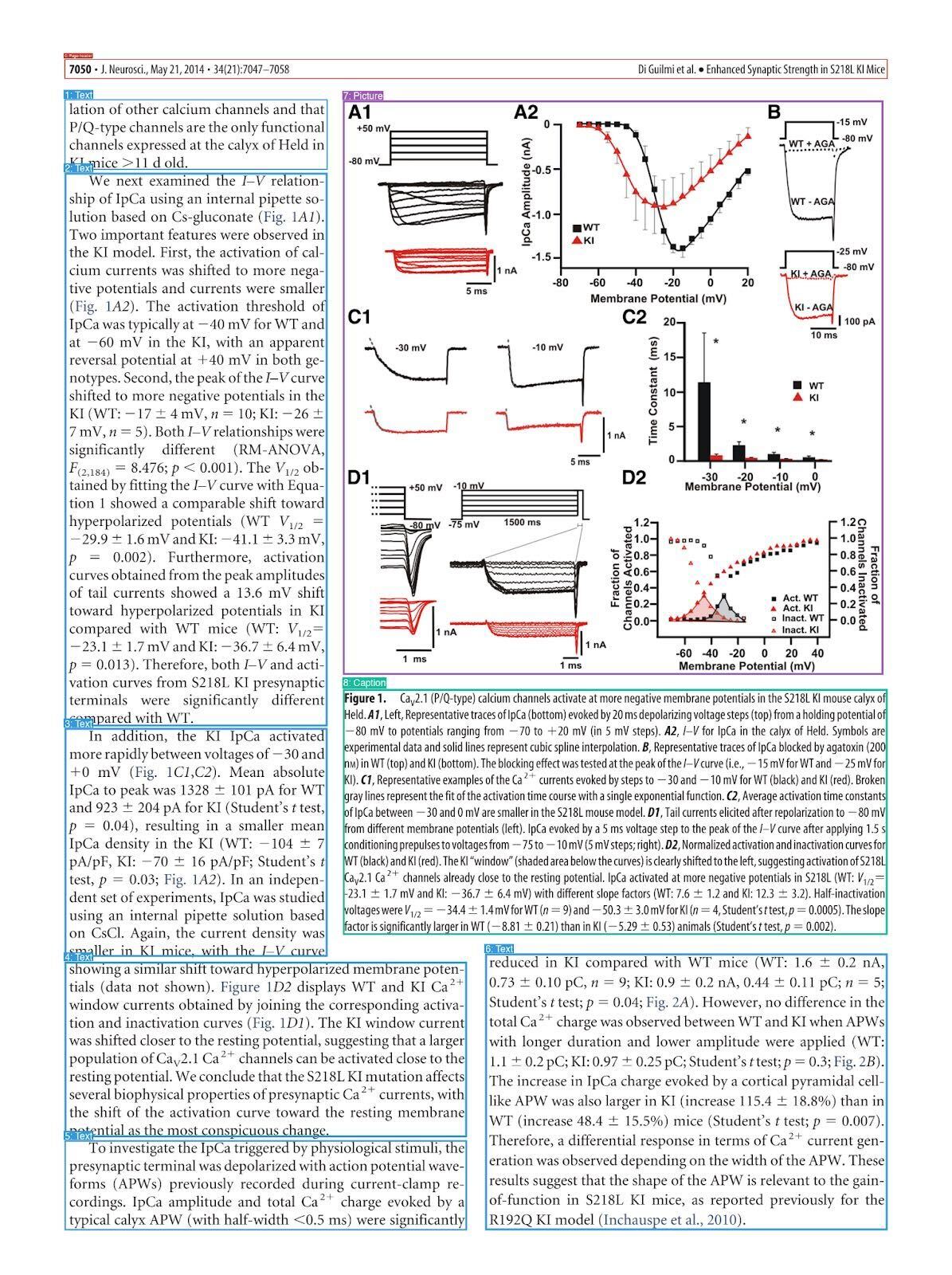
Figure 1 is correctly segmented to be a Picture, as shown in the purple bounding box.
Now the PDF has been successfully parsed, the next step is preparing for retrieval augmented generation. As PaperQA currently uses text embeddings, any noteworthy features of media such as figures or tables won’t shift the embedding. Thus, an enrichment step is performed using a mid-sized VLM to produce a detailed caption as shown below. Figure A2 is well detailed, mentioning identifying information such as color and basic trends in the line plot.
The media contains scientific figures (Figure 1 and subfigures A1, A2, B, C1, C2, D1, and D2) representing electrophysiological data and biophysical properties of presynaptic calcium currents (IpCa) recorded from the calyx of Held in wild-type (WT, black) and mutant (S218L KI, red) mice models.
- **Figure A1:** <truncated>
- **Figure A2:** The current-voltage (I-V) relationship with IpCa amplitude plotted against membrane potential. WT data (black) and KI data (red) demonstrate a leftward shift in the activation of IpCa in KI, indicating that mutant channels activate at more negative potentials.
- <truncated>
After parsing and embedding, the core of Literature commences, called ranked contextual summarization. This process involves the agent LLM proposing a question, such as the following:
In Di Guilmi et al. 2014 (S218L Cav2.1 knock-in mice; calyx of Held), at what membrane potential is the presynaptic calcium current amplitude comparable between WT and KI (from the I–V relationship)? Please extract the voltage point(s) where WT and KI currents are similar or cross.
Given this proposed question, PaperQA performs an embedding-based top-K retrieval of similar chunks. This retrieval also fetches any media paired with the chunk. PaperQA then uses a separate LLM to summarize the question’s relation to the retrieved chunk/media, including a relevance score on a scale of 1-10. Notably, Nemotron Parse’s crisp bounding boxes are important because bad crops become context rot for the contextual summarization process. Below is the generated contextual summary, which is rated 8/10. We can see through this summary, Literature has found Figure 1A2 to be relevant:
The requested crossing/similar-amplitude voltage point is not explicitly stated in the text, but Figure 1A2 (provided image) shows WT (black squares) and KI (red triangles) I–V curves approaching each other and appearing most similar/crossing near the left side where currents are close to 0, <truncated> . The image also suggests KI may be larger (more negative) than WT around -45 to -35 mV, whereas WT is larger around -20 to -10 mV, implying an intersection roughly in the -30 mV range where the curves are similar.
In the next gather evidence call, the agent further investigates with a newly-informed question:
From Figure 1A2 (IpCa I–V curve) in Di Guilmi et al. 2014 J Neurosci, estimate the membrane potential where WT (black squares) and S218L KI (red triangles) presynaptic calcium current amplitudes are approximately equal (i.e., intersection/crossover of I–V curves).
The correct answer is now present in this next contextual summary with a score of 9/10:
The attached figure includes panel A2 showing the presynaptic calcium current (IpCa) I–V relationship for WT (black squares) vs S218L KI (red triangles). The curves appear to cross where the current amplitudes are approximately equal, visually around the mid-negative voltage range near about -30 mV (roughly between -35 and -25 mV by eye). <truncated>.
From there, the literature planning shows how this is understood and correctly answers -30 mV.
Figure 1A2 shows the I–V relationship; visual inspection indicates the two curves cross near −30 mV, which is the non-trivial voltage where amplitudes are comparable.
Serving infrastructure
Nemotron Parse 1.1 is a VLM with 900M parameters, with open-weights available on Hugging Face. The model is served using vLLM and deployed by Edison through Modal Labs on NVIDIA H200 GPUs. Using Python 3.13.0, vLLM version 0.14.1’s precompiled wheels, Modal GPU snapshotting, and a base image of nvidia/cuda:12.9.0-devel-ubuntu24.04, a container’s cold start time oscillates between one and two minutes.
When benchmarking Literature with Nemotron Parse, 34 questions were executed concurrently. At this concurrency level, between 20 and 60 pages per second were submitted to Modal. Edison targeted a maximum latency of 10 seconds per page request. To meet this requirement, a target concurrency of 32 requests per container was configured with a completion token limit of 5,000. vLLM uses its fork of Flash Attention 2 by default for Nemotron Parse. Additional optimization using NVIDIA Dynamo-Triton Inference Server and vLLM hyperparameter tuning is expected to further increase allowable concurrency.
During benchmark runs, Modal dynamically provisioned between 20 and 40 containers to meet workload demand, with 99.9 percent of requests experiencing queue times under 2 seconds. Edison determined that this throughput is sufficient to support production workloads, including both programmatic usage by Kosmos agents and direct human interaction through the platform.
Downstream performance
To contextualize performance, Literature is compared against a frontier laboratory’s deep research agent configured with high reasoning effort, tool use enabled, and a per-question timeout of three hours.
This comparison evaluates Literature’s end-to-end effectiveness under realistic research conditions.
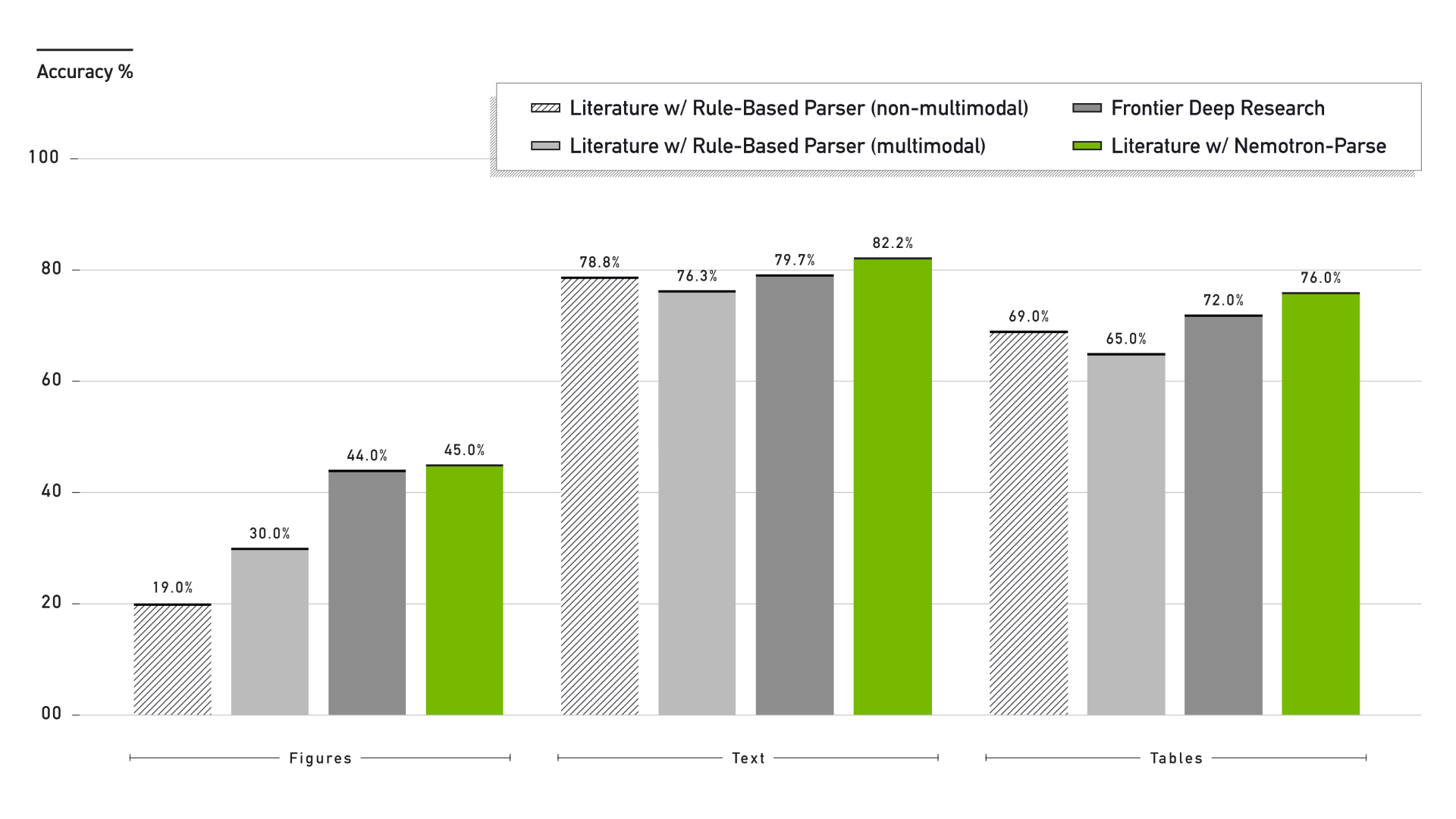
Many interesting observations can be made from Figure 3 above.
For the Figures task (FigQA2), transitioning from text-only processing to multimodal inference yields a substantial performance improvement. The text-only, rule-based parser achieves 19% accuracy, indicating that a subset of FigQA2 questions can be answered using textual references alone. However, Literature using Nemotron Parse outperforms the multimodal rule-based parser by 15 percentage points. This improvement can be attributed to two factors: higher recall of figure regions from PDFs and tighter bounding boxes around figures, which improve the quality and relevance of visual context provided to downstream models. The deep research system from a frontier laboratory also exceeds 19% accuracy on FigQA2, indicating that its internals utilize multimodal inference.
Despite these gains, FigQA2 accuracy remains below 50%. One contributing factor is document retrieval failure: in 16.4% of incorrect cases, the target PDF was not successfully retrieved due to missing search results or download failures. Additionally, multimodal reasoning remains an open challenge for large language models. Even when provided with correctly cropped figures, models may fail to interpret visual content accurately. The original LAB-Bench paper, using frontier models from summer 2024, reports zero-shot figure reasoning accuracy between 25% and 45%. LABBench2, now with winter 2025’s frontier models, similarly reports accuracy in the range of 45% to 65% showing models still struggle with figure inference. Smaller contributing factors include residual benchmark noise, occasional API timeouts, and rare cases where Nemotron Parse fails to generate an appropriate bounding box for edge-case figures.
For the text (LitQA3) and tables (TableQA2) inference tasks, introducing multimodality degrades performance when using a rule-based parser. This decline is indicative of context degradation caused by low-quality media extraction. In contrast, Literature using Nemotron Parse shows improved performance across both tasks, demonstrating that high-fidelity visual extraction can enrich context rather than dilute it. Although not shown in Figure 3, ablation studies using a text-only configuration of Nemotron Parse further indicate improved table understanding relative to the text-only rule-based parser. Across all three LABBench2 subsets, replacing a rule-based PDF parser with the model-based Nemotron Parse—without modifying any other system components—enables Literature to achieve state-of-the-art performance on LABBench2. These performance gains are now natively available in the PaperQA3 release.
Outcomes
Compared with the previously-used rule-based PDF parser, Nemotron Parse improves figure understanding by 15%, text understanding by 3%, and table understanding by 7% on the challenging LABBench2 benchmark.
By utilizing the < 1B parameter Nemotron Parse model, Literature now supports 100+ concurrent production runs with parallelized page processing, allowing a 300-page thesis to parse as quickly as 30-page papers.
Performing with a high reliability of PDF parsing, as demonstrated by LABBench2 performance surpassing a modern-day frontier lab’s deep research tool.
Mathematical formula understanding has vastly increased, as Nemotron Parse accurately converts equations to LaTeX.
“PDF extraction was a much larger bottleneck than initially realized...Switching to Nemotron Parse was a game changer, it unlocked the whole multimodal pipeline.”
James Braza
AI Research Technical Staff at Edison Scientific
Lessons Learned
Strategies for success
Evaluate early: Benchmarks showed that Nemotron Parse outperformed the most popular parsing alternatives, prompting Edison to test further. The team was also drawn to Nemotron Parse’s ability to output tables in LaTeX instead of Markdown, allowing more accurate representation of complex structures.
Leverage Nemotron Parse content classification: Edison’s pipeline takes different paths for each content type, for example utilizing the LaTeX representation of tables, but not the text export of figures.
Perform image enrichment: Use a VLM to augment the embedded text via an enrichment prompt. This enhances the embedding space to prevent retrieval from missing the image.
Implement failover routes: As of Nemotron Parse 1.1, some PDFs still cannot be parsed by the model. Failures usually manifest as hitting context limits or as the model proposing an invalid bounding box (e.g., a lower edge above an upper edge). In this case, you should develop failovers to rule-based PDF parsers, to ensure at least some content is parsed.
Tips for teams building similar systems
While Nemotron Parse may not be ideal for every use case, its superior accuracy and higher throughput compared with rule-based parsers makes it a better default choice.
Nemotron Parse supports three modes: text-only, bounding box-only, and text plus bounding box. Choose the modes that best suit your use cases.
Nemotron Parse implements image preprocessing logic to convert raw input images to the expected format.
A single test PDF is insufficient, so test with diverse real-world PDFs. Consider leveraging documents associated with open benchmarks like LAB-Bench to assess performance over a wide range of documents.
Related tutorials and resources
For developers who need highly capable scientific literature search and for those building their own scientific agents, the following resources provide step-by-step guidance and deeper technical detail:
How to Build a Document Processing Pipeline for RAG
NVIDIA Nemotron
Step-by-step instructions on how to set up a multimodal intelligent document processing pipeline using RAG.
Nemotron Parse
NVIDIA Nemotron
Visit the documentation for an overview of how to use the specialized VLM Nemotron Parse.
Use the Nemotron Parse Model
NVIDIA Nemotron
Available on Hugging Face or as an NVIDIA NIM™ API on build.nvidia.com.
Watch a Tutorial on Building a Document Processing Pipeline
NVIDIA Nemotron
Live technical walkthrough on using Nemotron Parse to process complex PDFs using RAG.
Build with the NVIDIA Blueprint for Enterprise RAG
NVIDIA Blueprint
Deploy an Enterprise RAG pipeline with accelerated microservices.
Kosmos by Edison Scientific
Edison Scientific Product
Accelerate breakthroughs with Kosmos, an AI-powered research platform that automates complex data analysis and literature synthesis to condense months of scientific work into a single day.
Acknowledgements
Many thanks to the teams at Edison Scientific, NVIDIA, and Modal—especially James Braza and Andrew White at Edison Scientific; Huiwen Ju, Xin Yu, Daniel Burkhardt, and Eliot Eshelman at NVIDIA; and Ben Shababo at Modal—for their help in writing and reviewing this case study.